2ET2
 
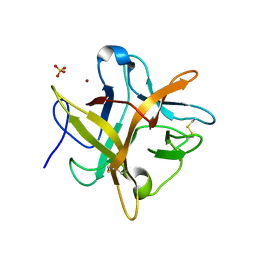 | | Crystal structure of an Asn to Ala mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
1Q2E
 
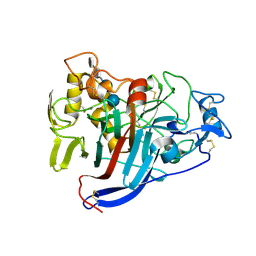 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
1X0S
 
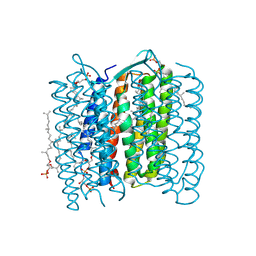 | | Crystal structure of the 13-cis isomer of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nishikawa, T, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the 13-cis isomer of bacteriorhodopsin in the dark-adapted state.
J.Mol.Biol., 352, 2005
|
|
2CIM
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
1Q8N
 
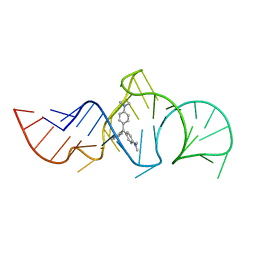 | | Solution Structure of the Malachite Green RNA Binding Aptamer | | Descriptor: | MALACHITE GREEN, RNA Aptamer | | Authors: | Flinders, J, DeFina, S.C, Brackett, D.M, Baugh, C, Wilson, C, Dieckmann, T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of planar and nonplanar ligands in the malachite green-RNA aptamer complex.
Chembiochem, 5, 2004
|
|
2CHH
 
 | | RALSTONIA SOLANACEARUM HIGH-AFFINITY MANNOSE-BINDING LECTIN | | Descriptor: | CALCIUM ION, PROTEIN RSC3288, UNKNOWN ATOM OR ION, ... | | Authors: | Mitchell, E.P, Wimmerova, M, Imberty, A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A new Ralstonia solanacearum high-affinity mannose-binding lectin RS-IIL structurally resembling the Pseudomonas aeruginosa fucose-specific lectin PA-IIL.
Mol. Microbiol., 52, 2004
|
|
1UCA
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1X0I
 
 | | Crystal Structure of the Acid Blue Form of Bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Okumura, H, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Acid Blue and Alkaline Purple Forms of Bacteriorhodopsin
J.Mol.Biol., 351, 2005
|
|
3U2U
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltotetraose | | Descriptor: | GLYCEROL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2ET8
 
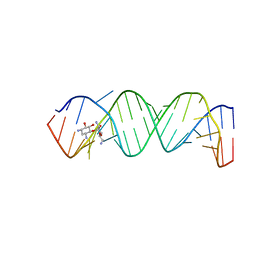 | | Complex Between Neamine and the 16S-RRNA A-Site | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Westhof, E. | | Deposit date: | 2005-10-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding.
Nucleic Acids Res., 33, 2005
|
|
1XG6
 
 | |
2FFR
 
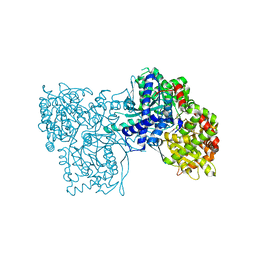 | | Crystallographic studies on N-azido-beta-D-glucopyranosylamine, an inhibitor of glycogen phosphorylase: comparison with N-acetyl-beta-D-glucopyranosylamine | | Descriptor: | Glycogen phosphorylase, muscle form, N-(azidoacetyl)-beta-D-glucopyranosylamine, ... | | Authors: | Petsalakis, E.I, Chrysina, E.D, Tiraidis, C, Hadjiloi, T, Leonidas, D.D, Oikonomakos, N.G, Aich, U, Varghese, B, Loganathan, D. | | Deposit date: | 2005-12-20 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic studies on N-azidoacetyl-beta-d-glucopyranosylamine, an inhibitor of glycogen phosphorylase: Comparison with N-acetyl-beta-d-glucopyranosylamine.
Bioorg.Med.Chem., 14, 2006
|
|
3AZZ
 
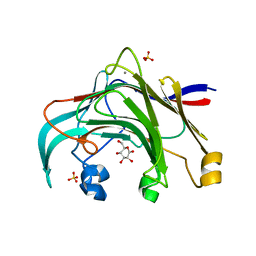 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with gluconolactone | | Descriptor: | CALCIUM ION, D-glucono-1,5-lactone, Laminarinase, ... | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
3B7L
 
 | | Human farnesyl diphosphate synthase complexed with MG and minodronate | | Descriptor: | (1-HYDROXY-2-IMIDAZO[1,2-A]PYRIDIN-3-YLETHANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Pilka, E.S, Dunford, J.E, Guo, K, Pike, A.C.W, Kavanagh, K.L, von Delft, F, Ebetino, F.H, Arrowsmith, C.H, Edwards, A.M, Russell, R.G.G, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human farnesyl diphosphate synthase complexed with MG and minodronate.
TO BE PUBLISHED
|
|
2E51
 
 | | Crystal structure of basic winged bean lectin in complex with A blood group disaccharide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Generation of blood group specificity: new insights from structural studies on the complexes of A- and B-reactive saccharides with basic winged bean agglutinin.
Proteins, 68, 2007
|
|
3TOP
 
 | | Crystral Structure of the C-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltase-glucoamylase, intestinal | | Authors: | Shen, Y, Qin, X.H, Ren, L.M. | | Deposit date: | 2011-09-06 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Structural insight into substrate specificity of human intestinal maltase-glucoamylase
Protein Cell, 2, 2011
|
|
3B30
 
 | | Crystal Structure of F. graminearum TRI101 complexed with Ethyl Coenzyme A | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Ethyl Coenzyme A, Trichothecene 3-O-acetyltransferase | | Authors: | Garvey, G.S, Rayment, I. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
2F6D
 
 | | Structure of the complex of a glucoamylase from Saccharomycopsis fibuligera with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Glucoamylase GLU1, PHOSPHATE ION, ... | | Authors: | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2005-11-29 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
2JKP
 
 | | Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCOSIDASE (ALPHA-GLUCOSIDASE SUSB), CALCIUM ION, ... | | Authors: | Gloster, T.M, Turkenburg, J.P, Potts, J.R, Henrissat, B, Davies, G.J. | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Divergence of Catalytic Mechanism within a Glycosidase Family Provides Insight Into Evolution of Carbohydrate Metabolism by Human Gut Flora.
Chem.Biol., 15, 2008
|
|
2F4S
 
 | | A-site RNA in complex with neamine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2FBZ
 
 | | Heme-No complex in a bacterial Nitric Oxide Synthase | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE, ... | | Authors: | Pant, K, Crane, B.R. | | Deposit date: | 2005-12-10 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nitrosyl-heme structures of Bacillus subtilis nitric oxide synthase have implications for understanding substrate oxidation.
Biochemistry, 45, 2006
|
|
3X27
 
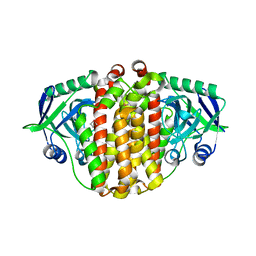 | | Structure of McbB in complex with tryptophan | | Descriptor: | Cucumopine synthase, TRYPTOPHAN | | Authors: | Mori, T, Sahashi, S, Morita, H, Abe, I. | | Deposit date: | 2014-12-10 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Structural Basis for beta-Carboline Alkaloid Production by the Microbial Homodimeric Enzyme McbB
Chem.Biol., 22, 2015
|
|
2K66
 
 | |
1EYL
 
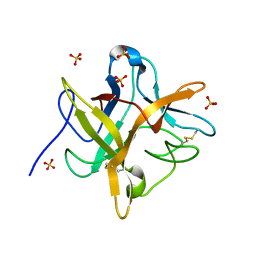 | | STRUCTURE OF A RECOMBINANT WINGED BEAN CHYMOTRYPSIN INHIBITOR | | Descriptor: | CHYMOTRYPSIN INHIBITOR, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Ghosh, S. | | Deposit date: | 2000-05-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
Protein Eng., 14, 2001
|
|
1EM1
 
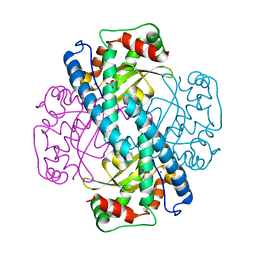 | | X-RAY CRYSTAL STRUCTURE FOR HUMAN MANGANESE SUPEROXIDE DISMUTASE, Q143A | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE, SULFATE ION | | Authors: | Leveque, V, Stroupe, M.E, Lepock, J.R, Cabelli, D.E, Tainer, J.A, Nick, H.S, Silverman, D.N. | | Deposit date: | 2000-03-14 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Multiple replacements of glutamine 143 in human manganese superoxide dismutase: effects on structure, stability, and catalysis.
Biochemistry, 39, 2000
|
|
