4ER4
 
 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
8CDB
 
 | | Proulilysin E229A structure | | Descriptor: | CALCIUM ION, Ulilysin, ZINC ION | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
4ER1
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, N-[(1R,2R,4R)-1-(cyclohexylmethyl)-2-hydroxy-6-methyl-4-{[(2R)-2-methylbutyl]carbamoyl}heptyl]-3-(1H-imidazol-3-ium-4-yl)-N~2~-[3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanoyl]-L-alaninamide | | Authors: | Quail, J.W, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-14 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
3MVH
 
 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | GSK3-beta peptide, MANGANESE (II) ION, N-{[(3S)-3-amino-1-(5-ethyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-yl]methyl}-2,4-difluorobenzamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3MV5
 
 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, GSK3-beta peptide, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-02 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
4ER2
 
 | | The active site of aspartic proteinases | | Descriptor: | ENDOTHIAPEPSIN, PEPSTATIN, SULFATE ION | | Authors: | Bailey, D, Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
3PRO
 
 | | ALPHA-LYTIC PROTEASE COMPLEXED WITH C-TERMINAL TRUNCATED PRO REGION | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ALPHA-LYTIC PROTEASE | | Authors: | Sauter, N.K, Mau, T, Rader, S.D, Agard, D.A. | | Deposit date: | 1998-08-26 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of alpha-lytic protease complexed with its pro region.
Nat.Struct.Biol., 5, 1998
|
|
4N7M
 
 | |
3KIV
 
 | | RECOMBINANT KRINGLE IV-10/M66 VARIANT OF HUMAN APOLIPOPROTEIN(A) | | Descriptor: | 6-AMINOHEXANOIC ACID, APOLIPOPROTEIN | | Authors: | Mochalkin, I, Tulinsky, A, Scanu, A. | | Deposit date: | 1998-09-08 | | Release date: | 1999-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recombinant kringle IV-10 modules of human apolipoprotein(a): structure, ligand binding modes, and biological relevance.
Biochemistry, 38, 1999
|
|
4N5D
 
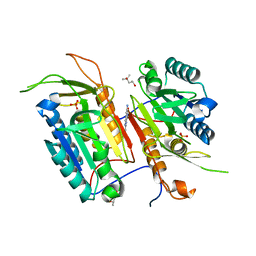 | | Tailoring Small Molecules for an Allosteric Site on Procaspase-6: Cpd1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-amino-2,8-dimethylpyrido[2,3-d]pyrimidin-7(8H)-one, Caspase-6, ... | | Authors: | Murray, J.M, Steffek, M. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tailoring small molecules for an allosteric site on procaspase-6.
Chemmedchem, 9, 2014
|
|
4NBN
 
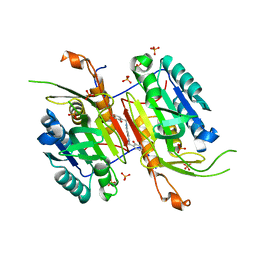 | |
4N7J
 
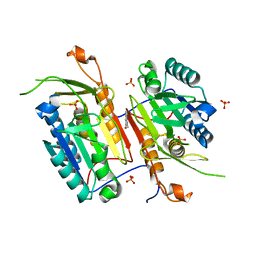 | |
4NBL
 
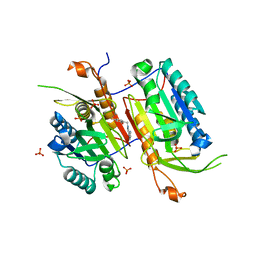 | |
4N6G
 
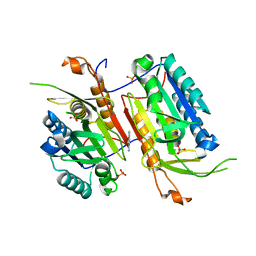 | |
4NBK
 
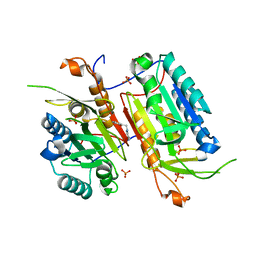 | |
4OUA
 
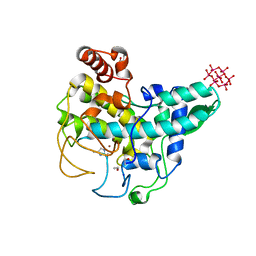 | | Coexistent single-crystal structure of latent and active mushroom tyrosinase (abPPO4) mediated by a hexatungstotellurate(VI) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-tungstotellurate(VI), COPPER (I) ION, ... | | Authors: | St.Mauracher, G, Molitor, C, Al-Oweini, R, Kortz, U, Rompel, A. | | Deposit date: | 2014-02-15 | | Release date: | 2014-06-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Latent and active abPPO4 mushroom tyrosinase cocrystallized with hexatungstotellurate(VI) in a single crystal.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3LQM
 
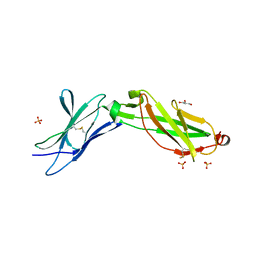 | | Structure of the IL-10R2 Common Chain | | Descriptor: | GLYCEROL, Interleukin-10 receptor subunit beta, SULFATE ION | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2010-02-09 | | Release date: | 2010-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and mechanism of receptor sharing by the IL-10R2 common chain.
Structure, 18, 2010
|
|
2WUB
 
 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp | | Descriptor: | FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, FAB FRAGMENT FAB40.DELTATRP LIGHT CHAIN, HEPATOCYTE GROWTH FACTOR ACTIVATOR LONG CHAIN, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
2V77
 
 | | Crystal Structure of Human Carboxypeptidase A1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBOXYPEPTIDASE A1, OCTANE-1,3,5,7-TETRACARBOXYLIC ACID, ... | | Authors: | Pallares, I, Fernandez, D, Comellas-Bigler, M, Fernandez-Recio, J, Ventura, S, Aviles, F.X, Vendrell, J. | | Deposit date: | 2007-07-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Direct interaction between a human digestive protease and the mucoadhesive poly(acrylic acid).
Acta Crystallogr. D Biol. Crystallogr., D64, 2008
|
|
3C9N
 
 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
4G6V
 
 | | CdiA-CT/CdiI toxin and immunity complex from Burkholderia pseudomallei | | Descriptor: | Adhesin/hemolysin, BROMIDE ION, CdiI | | Authors: | Morse, R.P, Nikolakakis, K, Willet, J, Gerrick, E, Low, D.A, Hayes, C.S, Goulding, C.W. | | Deposit date: | 2012-07-19 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1JQM
 
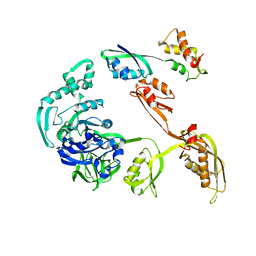 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQT
 
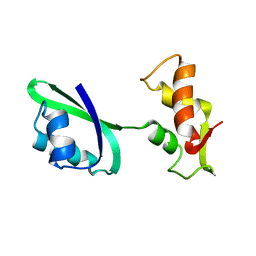 | | Fitting of L11 protein in the low resolution cryo-EM map of E.coli 70S ribosome | | Descriptor: | 50S Ribosomal protein L11 | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
4HAP
 
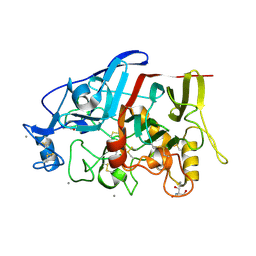 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HQO
 
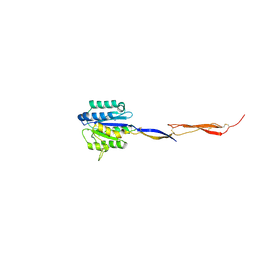 | | Crystal structure of Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
