6FGA
 
 | | Crystal structure of TRIM21 E3 ligase, RING domain in complex with its cognate E2 conjugating enzyme UBE2E1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, GLYCEROL, Ubiquitin-conjugating enzyme E2 E1, ... | | Authors: | Anandapadamanaban, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2018-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2.
J.Biol.Chem., 294, 2019
|
|
4AGH
 
 | | Structural features of ssDNA binding protein MoSub1 from Magnaporthe oryzae | | Descriptor: | MOSUB1, TRANSCRIPTION COFACTOR | | Authors: | Huang, J, Zhao, Y, Huang, D, liu, J, Peng, Y. | | Deposit date: | 2012-01-27 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Structural Features of the Single-Stranded DNA-Binding Protein Mosub1 from Magnaporthe Oryzae.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AMU
 
 | | Structure of ornithine carbamoyltransferase from Mycoplasma penetrans with a P321 space group | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | Gallego, P, Benach, J, Planell, R, Querol, E, Perez-Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
5FFS
 
 | | Crystal Structure of Surfactant Protein-A Y164A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
5XQU
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20I mutant (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3K7U
 
 | | Structure of essential protein from Trypanosoma brucei | | Descriptor: | Antibody, MP18 RNA editing complex protein | | Authors: | Wu, M. | | Deposit date: | 2009-10-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies.
J.Struct.Biol., 174, 2011
|
|
5D5A
 
 | | In meso in situ serial X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Liu, X, Kobilka, B, Kay Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4826 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FRK
 
 | | SeMet crystal structure of Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-18 | | Release date: | 2017-02-15 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
3QBU
 
 | | Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Shaik, M.M, Cendron, L, Percudani, R, Zanotti, G. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5701 Å) | | Cite: | The Structure of Helicobacter pylori HP0310 Reveals an Atypical Peptidoglycan Deacetylase.
Plos One, 6, 2011
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
5H59
 
 | | Ferredoxin-NADP+ reductase from maize root | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
6FIG
 
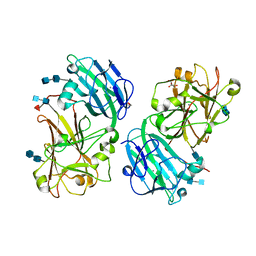 | | Crystal structure of the ANX1 ectodomain from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Santiago, J. | | Deposit date: | 2018-01-18 | | Release date: | 2018-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of two tandem malectin-like receptor kinases involved in plant reproduction.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3J7V
 
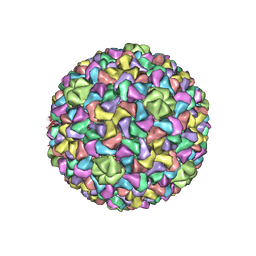 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4B6F
 
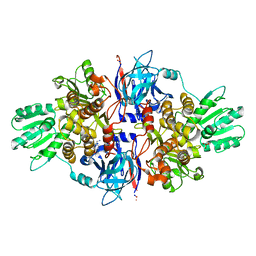 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (4-phenoxyphenyl)methylazanium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4B09
 
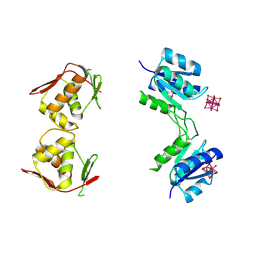 | | Structure of unphosphorylated BaeR dimer | | Descriptor: | HEXATANTALUM DODECABROMIDE, TRANSCRIPTIONAL REGULATORY PROTEIN BAER | | Authors: | Choudhury, H, Beis, K. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-10 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Dimeric Form of the Unphosphorylated Response Regulator Baer.
Protein Sci., 22, 2013
|
|
3AR9
 
 | | Calcium pump crystal structure with bound BeF3, TNP-AMP and TG in the absence of calcium | | Descriptor: | 2',3'-O-[(1r)-2,4,6-trinitrocyclohexa-2,5-diene-1,1-diyl]adenosine 5'-(dihydrogen phosphate), BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Toyoshima, C, Yonekura, S, Tsueda, J, Iwasawa, S. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AR3
 
 | | Calcium pump crystal structure with bound ADP and TG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Yonekura, S, Tsueda, J, Iwasawa, S. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3MYW
 
 | | The Bowman-Birk type inhibitor from mung bean in ternary complex with porcine trypsin | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Engh, R.A, Bode, W, Huber, R, Lin, G, Chi, C. | | Deposit date: | 2010-05-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 0.25-nm X-ray structure of the Bowman-Birk-type inhibitor from mung bean in ternary complex with porcine trypsin.
Eur.J.Biochem., 212, 1993
|
|
5GNI
 
 | |
5GZB
 
 | | Crystal Structure of Transcription Factor TEAD4 in Complex with M-CAT DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*TP*TP*CP*CP*TP*CP*TP*C)-3'), GLYCEROL, ... | | Authors: | He, F, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-04-19 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | DNA-binding mechanism of the Hippo pathway transcription factor TEAD4
Oncogene, 36, 2017
|
|
5H42
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
3AR7
 
 | | Calcium pump crystal structure with bound TNP-ATP and TG in the absence of Ca2+ | | Descriptor: | OCTANOIC ACID [3S-[3ALPHA, 3ABETA, 4ALPHA, ... | | Authors: | Toyoshima, C, Yonekura, S, Tsueda, J, Iwasawa, S. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AR5
 
 | | Calcium pump crystal structure with bound TNP-AMP and TG | | Descriptor: | 2',3'-O-[(1r)-2,4,6-trinitrocyclohexa-2,5-diene-1,1-diyl]adenosine 5'-(dihydrogen phosphate), OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Yonekura, S, Tsueda, J, Iwasawa, S. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5GRH
 
 | |
5CR2
 
 | |
