7M05
 
 | |
5MTV
 
 | | Active structure of EHD4 complexed with ATP-gamma-S | | Descriptor: | EH domain-containing protein 4, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Melo, A.A, Daumke, O. | | Deposit date: | 2017-01-10 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into the activation mechanism of dynamin-like EHD ATPases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5FTJ
 
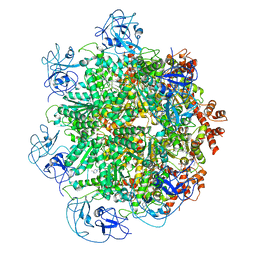 | | Cryo-EM structure of human p97 bound to UPCDC30245 inhibitor | | Descriptor: | 1-(3-(5-FLUORO-1H-INDOL-2-YL)PHENYL)PIPERIDIN-4-YL)(2-(4-ISOPROPYL-PIPERAZIN1-YL)ETHYL)-CARBAMATE, ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5MUN
 
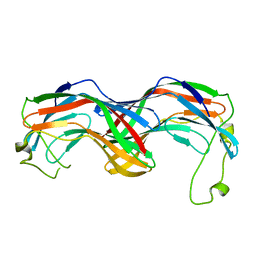 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
6C85
 
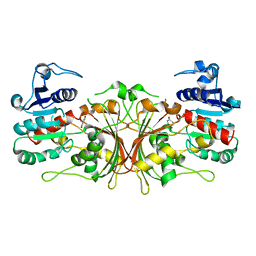 | |
5FVG
 
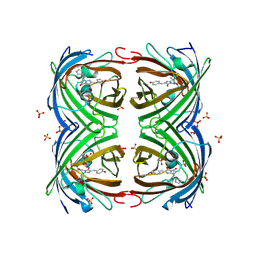 | | Structure of IrisFP at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett, 7, 2016
|
|
6C8F
 
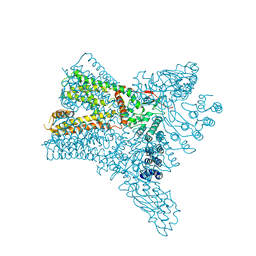 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of cesium | | Descriptor: | CESIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7OL9
 
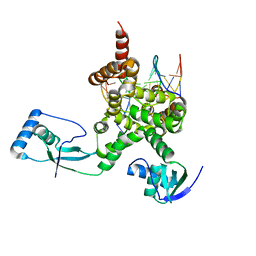 | |
1AIZ
 
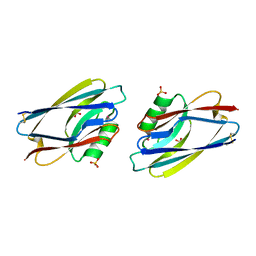 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, CADMIUM ION, SULFATE ION | | Authors: | Baker, E.N, Anderson, B.F, Blackwell, K.A. | | Deposit date: | 1993-11-11 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
7UJV
 
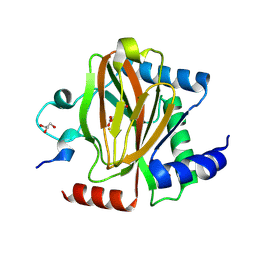 | | Structure of PHD2 in complex with HIF2a-CODD | | Descriptor: | Egl nine homolog 1, Endothelial PAS domain-containing protein 1, FE (III) ION, ... | | Authors: | Ferens, F.G, Tarade, D, Lee, J.E, Ohh, M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deficiency in PHD2-mediated hydroxylation of HIF2 alpha underlies Pacak-Zhuang syndrome.
Commun Biol, 7, 2024
|
|
5DN2
 
 | | Human NRP2 b1 domain in complex with the peptide corresponding to the C-terminus of VEGF-A | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Neuropilin-2, ... | | Authors: | Tsai, Y.C.I, Frankel, P, Fotinou, C, Rana, R, Zachary, I, Djordjevic, S. | | Deposit date: | 2015-09-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of neuropilin-2 reveal a zinc ion binding site remote from the vascular endothelial growth factor binding pocket.
Febs J., 283, 2016
|
|
1FW3
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PHOSPHOLIPASE A | | Authors: | Snijder, H.J, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-06-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of calcium binding and its role in activity and activation of outer membrane phospholipase A from Escherichia coli.
J.Mol.Biol., 309, 2001
|
|
5DOE
 
 | | Crystal structure of the Human Cytomegalovirus UL53 (residues 72-292) | | Descriptor: | Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
6JPB
 
 | | Rabbit Cav1.1-Diltiazem Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6C8G
 
 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of barium | | Descriptor: | BARIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6UMH
 
 | | Crystal structure of erenumab Fab-a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-PROPANDIOL, PHOSPHATE ION, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6C99
 
 | | Crystal structure of FcRn bound to UCB-303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Fox III, D, Abendroth, J, Porter, J, Deboves, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6JCT
 
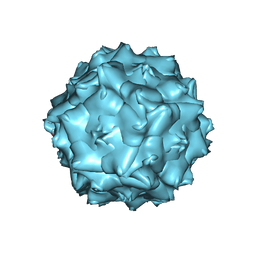 | | AAV5 in neutral condition at 3.18 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6TQL
 
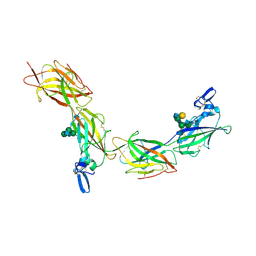 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
6TU4
 
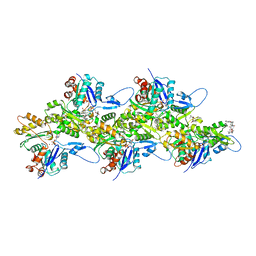 | | Structure of Plasmodium Actin1 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Vahokoski, J, Calder, L.J, Lopez, A.J, Rosenthal, P.B, Kursula, I. | | Deposit date: | 2020-01-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution structures of malaria parasite actomyosin and actin filaments.
Plos Pathog., 18, 2022
|
|
6CBK
 
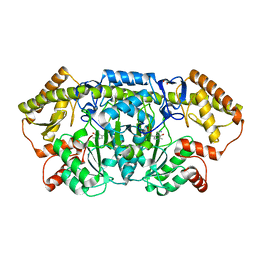 | | X-ray structure of NeoB from Streptomyces fradiae in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Neamine transaminase NeoN, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6BML
 
 | | Structure of human DHHC20 palmitoyltransferase, irreversibly inhibited by 2-bromopalmitate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PALMITIC ACID, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6BMU
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP244 | | Descriptor: | 4-[(2-chlorophenyl)methyl]-1-(2-hydroxy-3-methoxyphenyl)[1,2,4]triazolo[4,3-a]quinazolin-5(4H)-one, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6JPR
 
 | | Crystal structure of Phycocyanin from Nostoc sp. R76DM | | Descriptor: | GLYCEROL, PHYCOCYANOBILIN, Phycocyanin, ... | | Authors: | Sonani, R.R, Gupta, G.D, Rastogi, R.P, Patel, S.N, Madamwar, D, Kumar, V. | | Deposit date: | 2019-03-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phylogenetic and crystallographic analysis of Nostoc phycocyanin having blue-shifted spectral properties.
Sci Rep, 9, 2019
|
|
6BHD
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
