5EWJ
 
 | |
2BIU
 
 | | Crystal structure of human cyclophilin D at 1.7 A resolution, DMSO complex | | Descriptor: | DIMETHYL SULFOXIDE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5F0P
 
 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1(L557M) (SeMet labeled) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
3P4B
 
 | | Alternatingly modified 2'Fluoro RNA octamer f/rA2U2-P3 | | Descriptor: | 5'-R(*(CFZ)P*GP*(AF2)P*AP*(UFT)P*UP*(CFZ)P*G)-3', BARIUM ION | | Authors: | Pallan, P.S, Greene, E.M, Jicman, P.A, Pandey, R.K, Manoharan, M, Rozners, E, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
2BOX
 
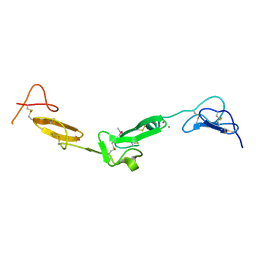 | | EGF Domains 1,2,5 of human EMR2, a 7-TM immune system molecule, in complex with strontium. | | Descriptor: | CACODYLATE ION, CHLORIDE ION, EGF-LIKE MODULE CONTAINING MUCIN-LIKE HORMONE RECEPTOR-LIKE 2 PRECURSOR, ... | | Authors: | Abbott, R.J.M, Spendlove, I, Roversi, P, Teriete, P, Knott, V, Handford, P.A, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2005-04-14 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of a Novel T Cell Receptor Co-Regulatory Protein Complex, Cd97-Cd55.
J.Biol.Chem., 282, 2007
|
|
3F2T
 
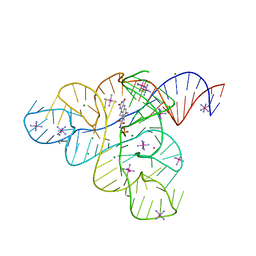 | |
6FFB
 
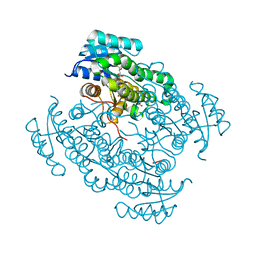 | | 17beta-hydroxysteroid dehydrogenase 14 variant S205 - mutant Q148A - in complex with a nonsteroidal inhibitor | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bertoletti, N, Heine, A, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2018-01-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational and structural studies uncover crucial amino acids determining activity and stability of 17 beta-HSD14.
J.Steroid Biochem.Mol.Biol., 189, 2019
|
|
1PZZ
 
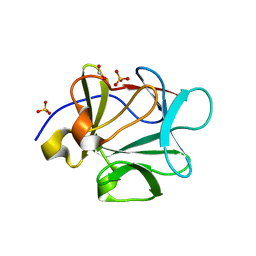 | | Crystal structure of FGF-1, V51N mutant | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-14 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
3SKK
 
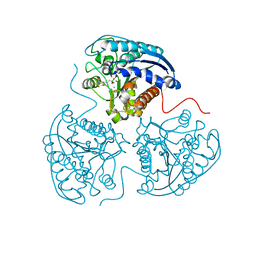 | | Crystal structure of human arginase I in complex with the inhibitor FABH, Resolution 1.70 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxy-6,6-difluorohexyl](trihydroxy)borate(1-) | | Authors: | Thorn, K.J, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
5GWO
 
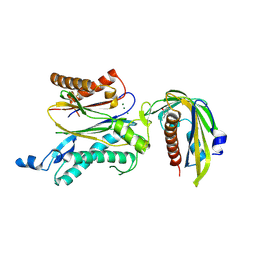 | | Crystal structure of RCAR3:PP2C S265F/I267M with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
6F69
 
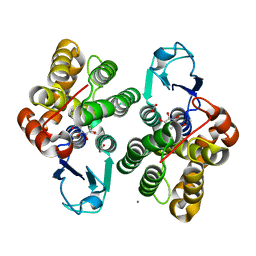 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with 2,3,4-Trihydroxybenzophenone | | Descriptor: | 2,3,4-Trihydroxybenzophenone, ACETATE ION, CALCIUM ION, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
3F65
 
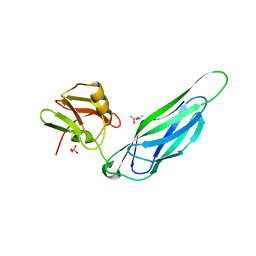 | | The F4 fimbrial chaperone FaeE does not self-cap its interactive surfaces | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chaperone protein faeE, PHOSPHATE ION | | Authors: | Van Molle, I, Moonens, K, Buts, L, Garcia-Pino, A, Wyns, L, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The F4 fimbrial chaperone FaeE is stable as a monomer that does not require self-capping of its pilin-interactive surfaces
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3WBJ
 
 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure II | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5CKE
 
 | | E.coli MazF E24A form IIa | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Endoribonuclease MazF, SULFATE ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
6F66
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with 2,4-Dihydroxybenzophenone | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
1NKW
 
 | | Crystal Structure Of The Large Ribosomal Subunit From Deinococcus Radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Harms, J.M, Schluenzen, F, Zarivach, R, Bashan, A, Gat, S, Agmon, I, Bartels, H, Franceschi, F, Yonath, A. | | Deposit date: | 2003-01-05 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High resolution structure of the large ribosomal subunit from a mesophilic eubacterium
Cell(Cambridge,Mass.), 107, 2001
|
|
3WN5
 
 | | Crystal structure of asymmetrically engineered Fc variant in complex with FcgRIIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kamikawa, T, Hattori, K. | | Deposit date: | 2013-12-05 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a novel asymmetrically engineered Fc variant with improved affinity for Fc gamma Rs.
Mol.Immunol., 58, 2014
|
|
3WVL
 
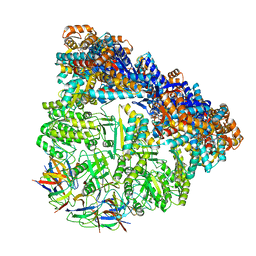 | | Crystal structure of the football-shaped GroEL-GroES complex (GroEL: GroES2:ATP14) from Escherichia coli | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koike-Takeshita, A, Arakawa, T, Taguchi, H, Shimamura, T. | | Deposit date: | 2014-05-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.788 Å) | | Cite: | Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 angstrom reveals rearrangement between two GroEL rings.
J.Mol.Biol., 426, 2014
|
|
3B9J
 
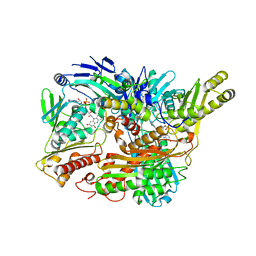 | | Structure of Xanthine Oxidase with 2-hydroxy-6-methylpurine | | Descriptor: | 6-methyl-3,9-dihydro-2H-purin-2-one, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Pauff, J.M, Zhang, J, Bell, C.E, Hille, R. | | Deposit date: | 2007-11-05 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate orientation in xanthine oxidase: crystal structure of enzyme in reaction with 2-hydroxy-6-methylpurine.
J.Biol.Chem., 283, 2008
|
|
5G43
 
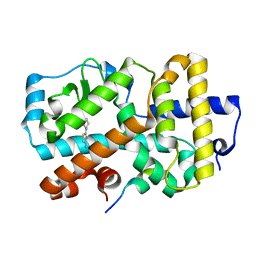 | | Ligand complex of RORg LBD | | Descriptor: | 2-(1-PIPERIDINYL)-1,3-THIAZOL-4-AMINE, NUCLEAR RECEPTOR ROR-GAMMA, RORG | | Authors: | Xue, Y, Guo, H, Hillertz, P. | | Deposit date: | 2016-05-04 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Fragment Screening of Rorgammat Using Cocktail Crystallography: Identification of Simultaneous Binding of Multiple Fragments.
Chemmedchem, 11, 2016
|
|
3WZY
 
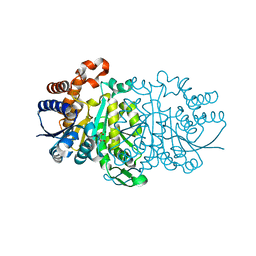 | |
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
5FTK
 
 | | Cryo-EM structure of human p97 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FQ8
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-decanoyloxypropyl decanoate, BT_2262 (UNCHARACTERISED LIPOPROTEIN), ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FYA
 
 | | Cubic crystal of the native PlpD | | Descriptor: | PATATIN-LIKE PROTEIN, PLPD | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
