3U0U
 
 | |
2ZE4
 
 | | Crystal structure of phospholipase D from streptomyces antibioticus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | Authors: | Suzuki, A, Kakuno, K, Saito, R, Iwasaki, Y, Yamane, T, Yamane, T. | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
3KO3
 
 | |
4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
5NZ8
 
 | | Clostridium thermocellum cellodextrin phosphorylase with cellotetraose and phosphate bound | | Descriptor: | Cellodextrin phosphorylase, PHOSPHATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | O'Neill, E.C, Pergolizzi, G, Stevenson, C.E.M, Lawson, D.M, Nepogodiev, S.A, Field, R.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cellodextrin phosphorylase from Ruminiclostridium thermocellum: X-ray crystal structure and substrate specificity analysis.
Carbohydr. Res., 451, 2017
|
|
3U08
 
 | | Crystal structure of DB1963-D(CGCGAATTCGCG)2 complex at 1.25 A resolution | | Descriptor: | 4'-(5-carbamimidoyl-1H-benzimidazol-2-yl)biphenyl-4-carboxamide, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Wei, D.G, Neidle, S. | | Deposit date: | 2011-09-28 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small-molecule binding to the DNA minor groove is mediated by a conserved water cluster.
J.Am.Chem.Soc., 135, 2013
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
1EEG
 
 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
5A6L
 
 | | High resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with two xylobiose units bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
2XGB
 
 | | Crystal structure of Barley Beta-Amylase complexed with 2,3- epoxypropyl-alpha-D-glucopyranoside | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
3AJ1
 
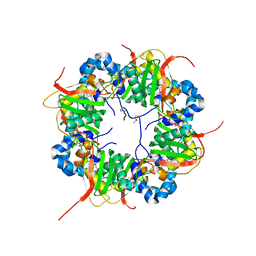 | | The structure of AxCeSD octamer (N-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4DVE
 
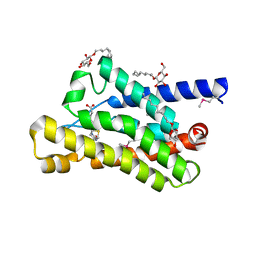 | | Crystal structure at 2.1 A of the S-component for biotin from an ECF-type ABC transporter | | Descriptor: | BIOTIN, Biotin transporter BioY, nonyl beta-D-glucopyranoside | | Authors: | Berntsson, R.P.-A, ter Beek, J, Majsnerowska, M, Duurkens, R, Puri, P, Poolman, B, Slotboom, D.J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural divergence of paralogous S components from ECF-type ABC transporters.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2Y5M
 
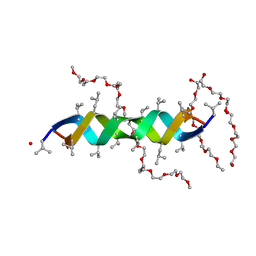 | |
6BOQ
 
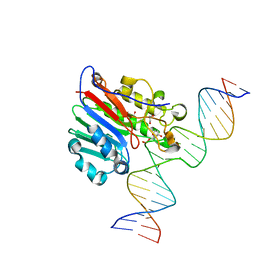 | | Human APE1 substrate complex with an A/A mismatch adjacent the THF | | Descriptor: | 1,2-ETHANEDIOL, 21-mer DNA, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4NQC
 
 | | Crystal structure of TCR-MR1 ternary complex and covalently bound 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T-cell activation by transitory neo-antigens derived from distinct microbial pathways.
Nature, 509, 2014
|
|
3PFP
 
 | | Structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis in complex with an active site inhibitor | | Descriptor: | (2R)-2,7-bis(phosphonooxy)heptanoic acid, (2S)-2,7-bis(phosphonooxy)heptanoic acid, CHLORIDE ION, ... | | Authors: | Reichau, S, Jiao, W, Walker, S.R, Hutton, R.D, Parker, E.J, Baker, E.N. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Potent inhibitors of a shikimate pathway enzyme from Mycobacterium tuberculosis: combining mechanism- and modeling-based design
J.Biol.Chem., 286, 2011
|
|
1A7Y
 
 | | CRYSTAL STRUCTURE OF ACTINOMYCIN D | | Descriptor: | ACTINOMYCIN D, ETHYL ACETATE, METHANOL | | Authors: | Schafer, M, Sheldrick, G.M, Bahner, I, Lackner, H. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
3UQ9
 
 | | Adenosine kinase from Schistosoma mansoni in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosine kinase, putative, ... | | Authors: | Romanello, L, Cassago, A, Bachega, F.R, Garatt, R.C, DeMarco, R, Pereira, H.M. | | Deposit date: | 2011-11-19 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Adenosine kinase from Schistosoma mansoni: structural basis for the differential incorporation of nucleoside analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1AE8
 
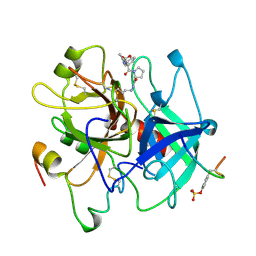 | | HUMAN ALPHA-THROMBIN INHIBITION BY EOC-D-PHE-PRO-AZALYS-ONP | | Descriptor: | 1-ETHOXYCARBONYL-D-PHE-PRO-2(4-AMINOBUTYL)HYDRAZINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
2WGU
 
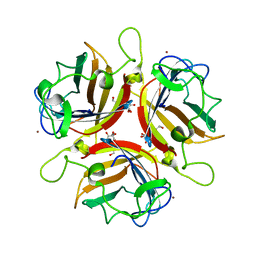 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N- methoxycarbonyl -3,5-dideoxy- D-glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-[(methoxycarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
4NZ1
 
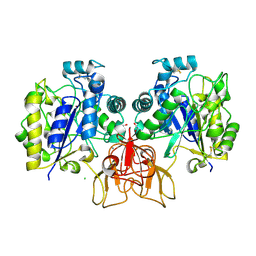 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
181D
 
 | |
180D
 
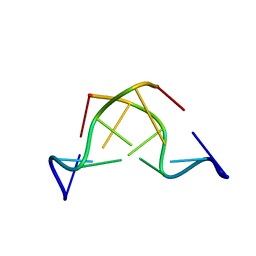 | |
1ABF
 
 | |
