2Z36
 
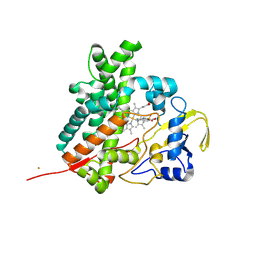 | | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 type compactin 3'',4''-hydroxylase, FE (III) ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Fujii, T, Arisawa, A, Tamura, T. | | Deposit date: | 2007-06-02 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105)
Biochem.Biophys.Res.Commun., 361, 2007
|
|
6JZJ
 
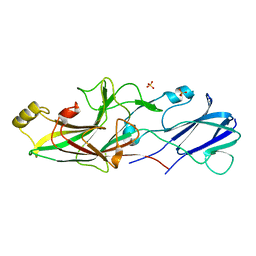 | | Structure of FimA type-2 (FimA2) prepilin of the type V major fimbrium | | Descriptor: | Major fimbrium subunit FimA type-2, SULFATE ION | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
2Z1H
 
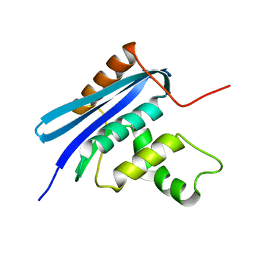 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
4OID
 
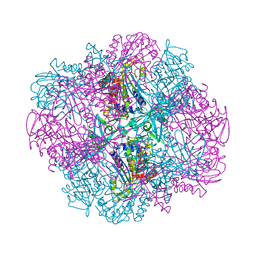 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | Probable M18 family aminopeptidase 2 | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
2Z23
 
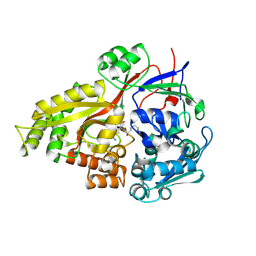 | | Crystal structure of Y.pestis oligo peptide binding protein OppA with tri-lysine ligand | | Descriptor: | Periplasmic oligopeptide-binding protein, peptide (LYS)(LYS)(LYS) | | Authors: | Tanabe, M, Bertland, T, Mirza, O, Byrne, B, Brown, K.A. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2Z3O
 
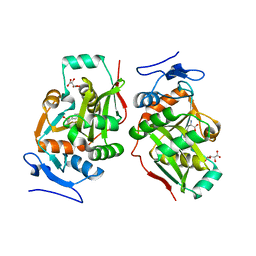 | |
3V0X
 
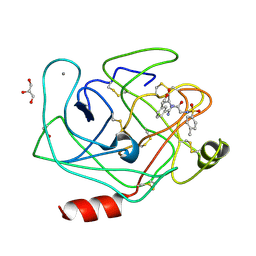 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bovine trypsin variants X(tripleGlu217Phe227) in complexes with small molecule inhibitor
To be Published
|
|
3V3T
 
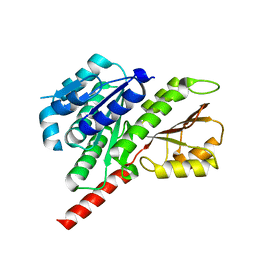 | |
6JZE
 
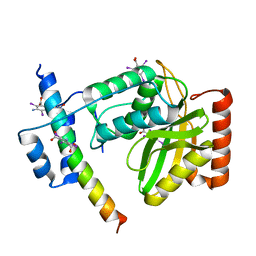 | | Crystal structure of VASH2-SVBP complex with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | Deposit date: | 2019-05-01 | | Release date: | 2019-08-07 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
4OFQ
 
 | | Structure of the C-terminal domain of the Streptococcus pyogenes antigen I/II-family protein AspA | | Descriptor: | CALCIUM ION, Putative cell surface protein | | Authors: | Hall, M, Nylander, A, Jenkinson, H.F, Persson, K. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of AspA (antigen I/II-family) protein from Streptococcus pyogenes.
FEBS Open Bio, 4, 2014
|
|
3V4O
 
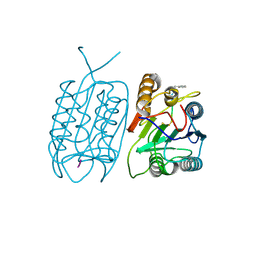 | |
4OJZ
 
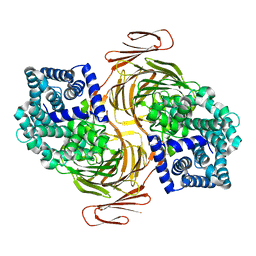 | |
2Z5P
 
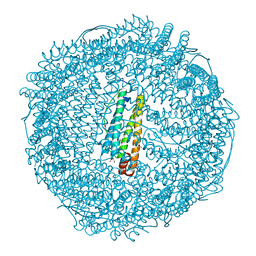 | | Apo-Fr with low content of Pd ions | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Ueno, T, Hirata, K, Abe, M, Suzuki, M, Abe, S, Shimizu, N, Yamamoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Process of accumulation of metal ions on the interior surface of apo-ferritin: crystal structures of a series of apo-ferritins containing variable quantities of Pd(II) ions.
J.Am.Chem.Soc., 131, 2009
|
|
2Z69
 
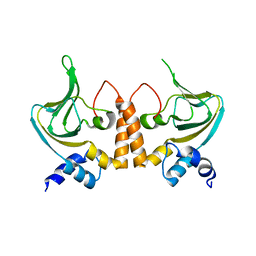 | |
3V1Z
 
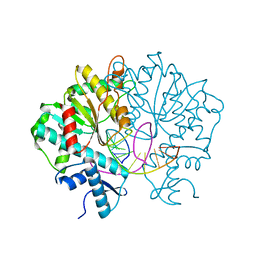 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP*G)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
4OLW
 
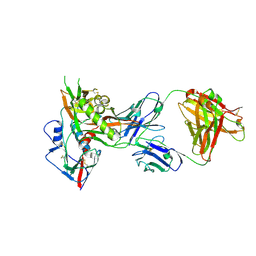 | |
4OGY
 
 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
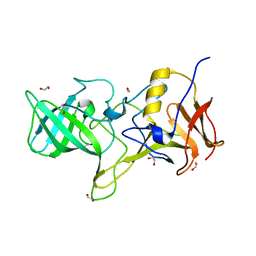
4OOS
 
 | |
6K39
 
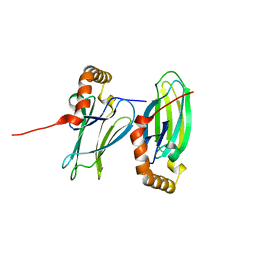 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3981427 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
4OMF
 
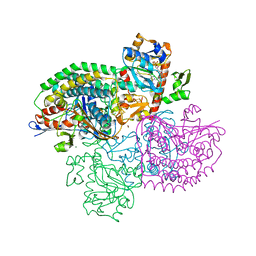 | | The F420-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member | | Descriptor: | CHLORIDE ION, F420-reducing hydrogenase, subunit alpha, ... | | Authors: | Vitt, S, Ma, K, Warkentin, E, Moll, J, Pierik, A, Shima, S, Ermler, U. | | Deposit date: | 2014-01-27 | | Release date: | 2014-06-11 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The F420-Reducing [NiFe]-Hydrogenase Complex from Methanothermobacter marburgensis, the First X-ray Structure of a Group 3 Family Member.
J.Mol.Biol., 426, 2014
|
|
3UOZ
 
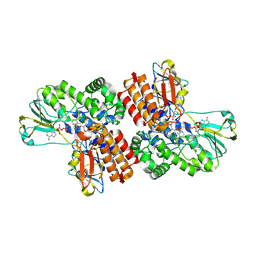 | | Crystal Structure of OTEMO complex with FAD and NADP (form 2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
6K67
 
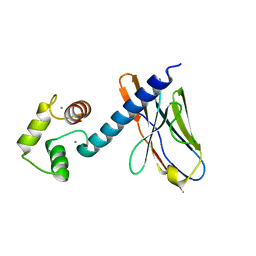 | | Application of anti-helix antibodies in protein structure determination (9011-3LRH) | | Descriptor: | 3LRH introbody, CALCIUM ION, Engineered calmodulin | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4OS4
 
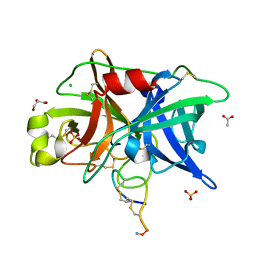 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK603 (bicyclic 1) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OSH
 
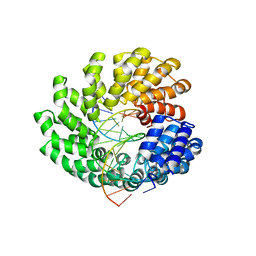 | | Crystal structure of the TAL effector dHax3 with NI RVD at 2.2 angstrom resolution | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
6K7V
 
 | | Structure of NLRP1 CARD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Gong, Q, Xu, C, Zhang, J, Wu, B. | | Deposit date: | 2019-06-09 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
