1RQT
 
 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQU
 
 | | NMR structure of L7 dimer from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQV
 
 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
1RQW
 
 | |
1RQX
 
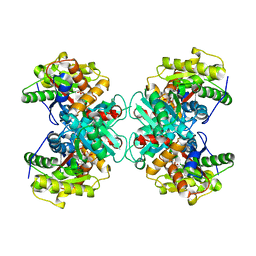 | | Crystal structure of ACC Deaminase complexed with Inhibitor | | Descriptor: | 1-AMINOCYCLOPROPYLPHOSPHONATE, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhao, Z, Kao, C.L, Zhou, Q, Tao, Z, Zhang, H, Liu, H.W. | | Deposit date: | 2003-12-07 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of 1-aminocyclopropane-1-carboxylate deaminase: observation of an aminyl intermediate and identification of Tyr 294 as the active-site nucleophile.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1RQY
 
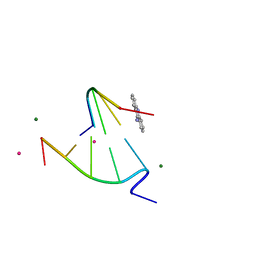 | | 9-amino-[N-(2-dimethylamino)proply]-acridine-4-carboxamide bound to d(CGTACG)2 | | Descriptor: | 5'-D(CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[3-(DIMETHYLAMINO)PROPYL]ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, ... | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P. | | Deposit date: | 2003-12-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 9-amino-[N-(2-dimethylamino)propyl]acridine-4-carboxamide bound to d(CGTACG)(2): a comparison of structures of d(CGTACG)(2) complexed with intercalatorsin the presence of cobalt.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RR2
 
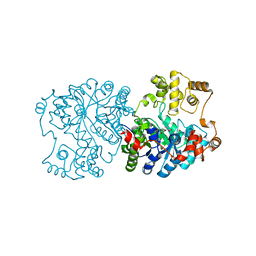 | | Propionibacterium shermanii transcarboxylase 5S subunit bound to 2-ketobutyric acid | | Descriptor: | 2-KETOBUTYRIC ACID, COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-08 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1RR6
 
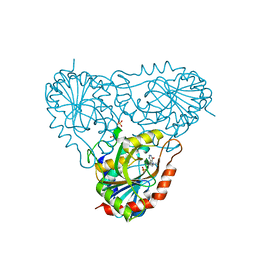 | | Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and phosphate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-12-08 | | Release date: | 2005-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasmodium falciparum purine nucleoside phosphorylase: crystal structures, immucillin inhibitors, and dual catalytic function.
J.Biol.Chem., 279, 2004
|
|
1RR7
 
 | |
1RR8
 
 | | Structural Mechanisms of Camptothecin Resistance by Mutations in Human Topoisomerase I | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T*GP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Chrencik, J.E, Staker, B.L, Burgin, A.B, Stewart, L, Redinbo, M.R. | | Deposit date: | 2003-12-08 | | Release date: | 2004-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of camptothecin resistance by human topoisomerase I mutations
J.Mol.Biol., 339, 2004
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1RRA
 
 | | RIBONUCLEASE A FROM RATTUS NORVEGICUS (COMMON RAT) | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBONUCLEASE) | | Authors: | Gupta, V, Muyldermans, S, Wyns, L, Salunke, D. | | Deposit date: | 1998-12-04 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of recombinant rat pancreatic RNase A.
Proteins, 35, 1999
|
|
1RRB
 
 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
1RRC
 
 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-GTC-3' SSDNA | | Descriptor: | 5'-D(*GP*TP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1RRD
 
 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH RNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1RRE
 
 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1RRF
 
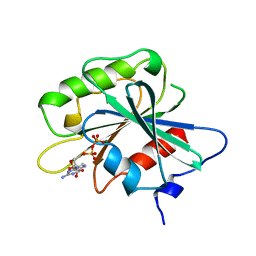 | | NON-MYRISTOYLATED RAT ADP-RIBOSYLATION FACTOR-1 COMPLEXED WITH GDP, MONOMERIC CRYSTAL FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAT ADP-RIBOSYLATION FACTOR-1 | | Authors: | Greasley, S.E, Jhoti, H, Bax, B. | | Deposit date: | 1995-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms.
Nat.Struct.Biol., 2, 1995
|
|
1RRG
 
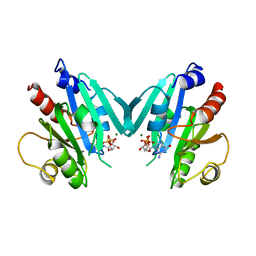 | | NON-MYRISTOYLATED RAT ADP-RIBOSYLATION FACTOR-1 COMPLEXED WITH GDP, DIMERIC CRYSTAL FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAT ADP-RIBOSYLATION FACTOR-1 | | Authors: | Greasley, S.E, Jhoti, H, Bax, B. | | Deposit date: | 1995-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms.
Nat.Struct.Biol., 2, 1995
|
|
1RRH
 
 | |
1RRI
 
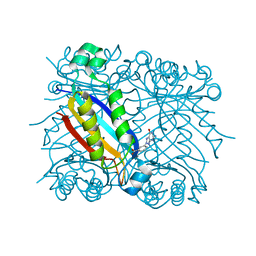 | | DHNA complex with 3-(5-amino-7-hydroxy-[1,2,3] triazolo [4,5-d]pyrimidin-2-yl)-benzoic acid | | Descriptor: | 3-(5-AMINO-7-HYDROXY-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-2-YL)-BENZOIC ACID, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
1RRJ
 
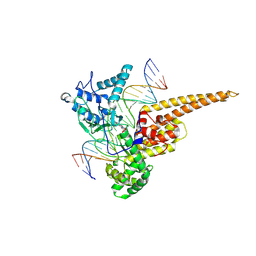 | | Structural Mechanisms of Camptothecin Resistance by Mutations in Human Topoisomerase I | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T*GP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Chrencik, J.E, Staker, B.L, Burgin, A.B, Stewart, L, Redinbo, M.R. | | Deposit date: | 2003-12-08 | | Release date: | 2004-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of camptothecin resistance by human topoisomerase I mutations
J.Mol.Biol., 339, 2004
|
|
1RRK
 
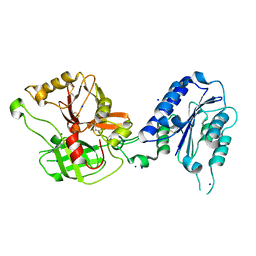 | | Crystal Structure Analysis of the Bb segment of Factor B | | Descriptor: | COBALT (II) ION, Complement factor B, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1RRL
 
 | |
1RRM
 
 | |
1RRO
 
 | | REFINEMENT OF RECOMBINANT ONCOMODULIN AT 1.30 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, RAT ONCOMODULIN | | Authors: | Ahmed, F.R, Rose, D.R, Evans, S.V, Pippy, M.E, To, R. | | Deposit date: | 1992-08-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refinement of recombinant oncomodulin at 1.30 A resolution.
J.Mol.Biol., 230, 1993
|
|
