7NYH
 
 | |
7NYU
 
 | |
7OC1
 
 | | Structure of Pseudomonas aeruginosa FabF mutant C164Q in complex with Platensimycin | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O, Klein, R. | | Deposit date: | 2021-04-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Experimental Toolbox for Structure-Based Hit Discovery for P. aeruginosa FabF, a Promising Target for Antibiotics.
Chemmedchem, 16, 2021
|
|
7O6N
 
 | | Crystal structure of C. elegans ERH-2 PID-3 complex | | Descriptor: | Enhancer of rudimentary homolog 2, FORMIC ACID, Protein pid-3 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7OCX
 
 | |
7NYR
 
 | |
6N8G
 
 | | IRAK4 bound to benzoxazole compound | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[2-(morpholin-4-yl)-6-(piperidin-1-yl)-1,3-benzoxazol-5-yl]-6-(1H-pyrrolo[2,3-b]pyridin-5-yl)pyridine-2-carboxamide | | Authors: | Larsen, N.A, Bloudoff, K, Subramanian, V, Dobrzanska, M, Gluza, K. | | Deposit date: | 2018-11-29 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | To be published
To Be Published
|
|
7OCZ
 
 | |
7O6L
 
 | | Crystal structure of C. elegans ERH-2 | | Descriptor: | Enhancer of rudimentary homolog 2 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7NYV
 
 | |
7OOA
 
 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6N9F
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic ACCA-DPhe and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
7OO0
 
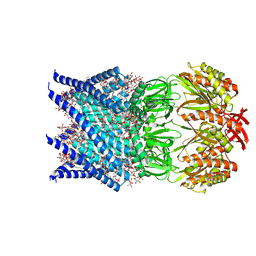 | | Mechanosensitive channel MscS solubilized with DDM in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel MscS | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ONJ
 
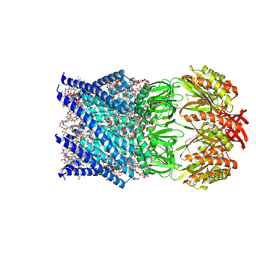 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO6
 
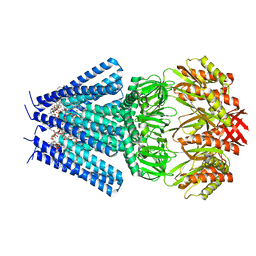 | | Mechanosensitive channel MscS solubilized with DDM in closed conformation with added lipid | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel of small conductance (MscS) | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO8
 
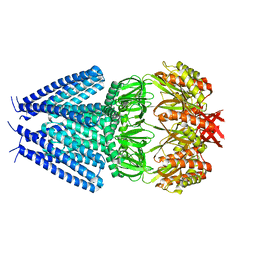 | |
7O1V
 
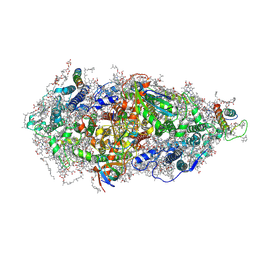 | | Structure of a Minimal Photosystem I | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Nelson, N, Caspy, I, Lambrev, P. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Two-Dimensional Electronic Spectroscopy of a Minimal Photosystem I Complex Reveals the Rate of Primary Charge Separation.
J.Am.Chem.Soc., 143, 2021
|
|
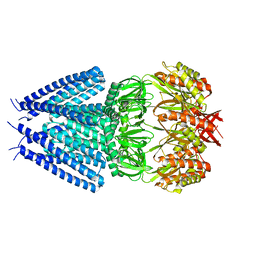
7ONL
 
 | |
6NFA
 
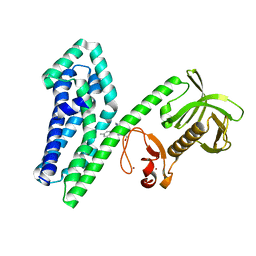 | |
3M8M
 
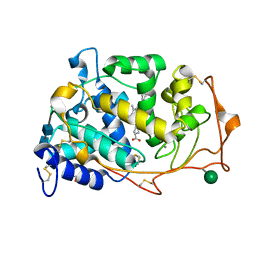 | | 1.05 A Structure of Manganese-free Manganese Peroxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Sundaramoorthy, M, Gold, M.H, Poulos, T.L. | | Deposit date: | 2010-03-18 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ultrahigh (0.93A) resolution structure of manganese peroxidase from Phanerochaete chrysosporium: implications for the catalytic mechanism.
J.Inorg.Biochem., 104, 2010
|
|
7NXL
 
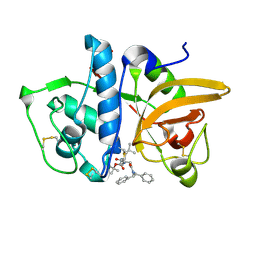 | | Structure of human cathepsin K in complex with the acrylamide inhibitor Gu3110 | | Descriptor: | Cathepsin K, SULFATE ION, tert-butyl (1-((4-(dibenzylamino)-4-oxobutyl)amino)-4-methyl-1-oxopentan-2-yl)carbamate | | Authors: | Busa, M, Benysek, J, Lemke, C, Gutschow, M, Mares, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Activity-Based Probe for Cathepsin K Imaging with Excellent Potency and Selectivity.
J.Med.Chem., 64, 2021
|
|
6SU0
 
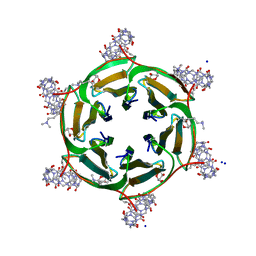 | | Crystal structure of dimethylated RSLex in complex with cucurbit[7]uril | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, cucurbit[7]uril, ... | | Authors: | Guagnini, F, Engilberge, S, Crowley, P.B. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineered assembly of a protein-cucurbituril biohybrid.
Chem.Commun.(Camb.), 56, 2020
|
|
7OKW
 
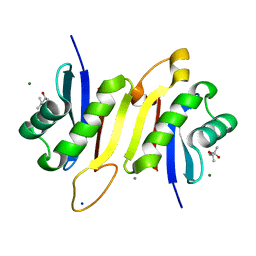 | | 1.62A X-ray crystal structure of the conserved C-terminal (CCT) of human OSR1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bax, B.D, Elvers, K.T, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
7O86
 
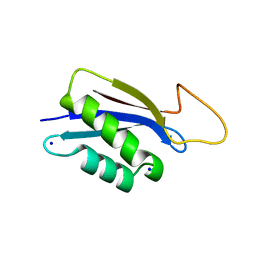 | | 1.73A X-ray crystal structure of the conserved C-terminal (CCT) of human SPAK | | Descriptor: | CALCIUM ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Elvers, K.T, Bax, B.D, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
7O7B
 
 | |
