2XJ4
 
 | |
4JQF
 
 | | Structure of the C-terminal domain of human telomeric Stn1 | | Descriptor: | CST complex subunit STN1 | | Authors: | Bryan, C.F, Rice, C.T, Harkisheimer, M, Schultz, D, Skordalakes, E. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-05 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human telomeric stn1-ten1 capping complex.
Plos One, 8, 2013
|
|
1MDI
 
 | |
1MDK
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN NFKB (RESIDUES 56-68 OF THE P50 SUBUNIT OF NFKB) | | Descriptor: | TARGET SITE IN HUMAN NFKB, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1995-02-27 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B.
Structure, 3, 1995
|
|
1MDJ
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN NFKB (RESIDUES 56-68 OF THE P50 SUBUNIT OF NFKB) | | Descriptor: | TARGET SITE IN HUMAN NFKB, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1995-02-27 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B.
Structure, 3, 1995
|
|
2QNL
 
 | |
1SQG
 
 | | The crystal structure of the E. coli Fmu apoenzyme at 1.65 A resolution | | Descriptor: | SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
3VOW
 
 | | Crystal Structure of the Human APOBEC3C having HIV-1 Vif-binding Interface | | Descriptor: | CHLORIDE ION, Probable DNA dC->dU-editing enzyme APOBEC-3C, ZINC ION | | Authors: | Kitamura, S, Suzuki, A, Watanabe, N, Iwatani, Y. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7ZCV
 
 | | Rgg144 of Streptococcus pneumoniae | | Descriptor: | Transcriptional regulator | | Authors: | Wallis, R, Girija, U.V, Yesilkaya, H. | | Deposit date: | 2022-03-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analysis for the development of peptide inhibitors for a Gram-positive quorum sensing system.
Mol.Microbiol., 117, 2022
|
|
7Z21
 
 | | BAF A12T bound to the lamin A/C Ig-fold domain | | Descriptor: | Barrier-to-autointegration factor, N-terminally processed, CHLORIDE ION, ... | | Authors: | Marcelot, A, Legrand, P, Zinn-Justin, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | The BAF A12T mutation disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures in Nestor-Guillermo progeria syndrome cells.
Nucleic Acids Res., 50, 2022
|
|
1N7B
 
 | | RIP-Radiation-damage Induced Phasing | | Descriptor: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | Authors: | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | Deposit date: | 2002-11-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
3VM8
 
 | |
1J74
 
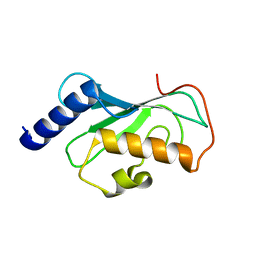 | | Crystal Structure of Mms2 | | Descriptor: | MMS2 | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pastushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
3VOX
 
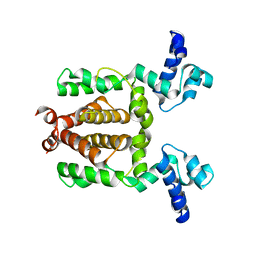 | |
3VP5
 
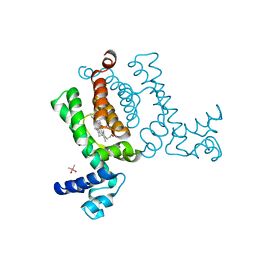 | | X-ray Crystal Structure of Wild Type HrtR in the Holo Form | | Descriptor: | CACODYLATE ION, PROTOPORPHYRIN IX CONTAINING FE, Transcriptional regulator | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Transcriptional Regulation of Heme Homeostasis in Lactococcus lactis.
J.Biol.Chem., 287, 2012
|
|
1JRN
 
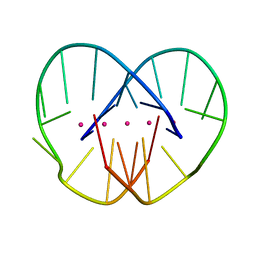 | |
7X3A
 
 | | NMR solution structure of the 1:1 complex of a pyridostatin (PDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-azanylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
7X8M
 
 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | Descriptor: | BERBERINE, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8O
 
 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
3IFV
 
 | | Crystal structure of the Haloferax volcanii proliferating cell nuclear antigen | | Descriptor: | PCNA, SODIUM ION | | Authors: | Winter, J.A, Christofi, P, Morroll, S, Bunting, K.A. | | Deposit date: | 2009-07-26 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Haloferax volcanii proliferating cell nuclear antigen reveals unique surface charge characteristics due to halophilic adaptation
Bmc Struct.Biol., 9, 2009
|
|
1OB9
 
 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
7ZMO
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-052 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-052, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMV
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMT
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMP
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-055 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-055, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.626 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
