3S76
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1H-imidazole-2-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S73
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2RAY
 
 | |
2RB1
 
 | |
3S71
 
 | |
3NX8
 
 | | human cAMP dependent protein kinase in complex with phenol | | Descriptor: | PHENOL, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-07-13 | | Release date: | 2011-07-13 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3L1B
 
 | | Complex Structure of FXR Ligand-binding domain with a tetrahydroazepinoindole compound | | Descriptor: | 1-methylethyl 8-fluoro-1,1-dimethyl-3-{[4-(3-morpholin-4-ylpropoxy)phenyl]carbonyl}-1,2,3,6-tetrahydroazepino[4,5-b]indole-5-carboxylate, Farnesoid X receptor | | Authors: | Xu, W, Lundquist, J.T. | | Deposit date: | 2009-12-11 | | Release date: | 2010-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Physiochemical Properties of the Tetrahydroazepinoindole Series of Farnesoid X Receptor (FXR) Agonists: Beneficial Modulation of Lipids in Primates.
J.Med.Chem., 53, 2010
|
|
3GS2
 
 | | Ring1B C-terminal domain/Cbx7 Cbox Complex | | Descriptor: | Chromobox protein homolog 7, E3 ubiquitin-protein ligase RING2, SULFATE ION, ... | | Authors: | Wang, R, Taylor, A.B, Kim, C.A. | | Deposit date: | 2009-03-26 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
8OOG
 
 | |
4XSM
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | Descriptor: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
2GCB
 
 | | G51S/S52T double mutant of L. casei FPGS | | Descriptor: | Folylpolyglutamate synthase | | Authors: | Smith, C.A, Cross, J.A, Bognar, A.L, Sun, X. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutation of Gly51 to serine in the P-loop of Lactobacillus casei folylpolyglutamate synthetase abolishes activity by altering the conformation of two adjacent loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3H57
 
 | | Myoglobin Cavity Mutant H64LV68N Deoxy form | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Soman, J, Olson, J.S. | | Deposit date: | 2009-04-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optical detection of disordered water within a protein cavity.
J.Am.Chem.Soc., 131, 2009
|
|
3D0X
 
 | |
3HCT
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3H8H
 
 | | Structure of the C-terminal domain of human RNF2/RING1B; | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RING2, GLYCEROL, ... | | Authors: | Walker, J.R, Bezsonova, I, Bacik, J, Duan, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins.
Biochemistry, 48, 2009
|
|
2GCA
 
 | | apo form of L. casei FPGS | | Descriptor: | Folylpolyglutamate synthase | | Authors: | Smith, C.A, Cross, J.A, Bognar, A.L, Sun, X. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutation of Gly51 to serine in the P-loop of Lactobacillus casei folylpolyglutamate synthetase abolishes activity by altering the conformation of two adjacent loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5C4U
 
 | |
1A97
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH GMP | | Descriptor: | BORIC ACID, GUANOSINE-5'-MONOPHOSPHATE, XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
8ESK
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, resting-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F6Y
 
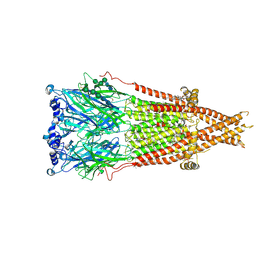 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with etomidate, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F6Z
 
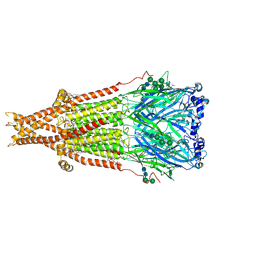 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with succinylcholine, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2,2'-[(1,4-DIOXOBUTANE-1,4-DIYL)BIS(OXY)]BIS(N,N,N-TRIMETHYLETHANAMINIUM), 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F2S
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, pore-blocked state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
5C4S
 
 | |
1Q0C
 
 | | Anerobic Substrate Complex of Homoprotocatechuate 2,3-Dioxygenase from Brevibacterium fuscum. (Complex with 3,4-Dihydroxyphenylacetate) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
7W1Q
 
 | |
