1SSA
 
 | | A STRUCTURAL INVESTIGATION OF CATALYTICALLY MODIFIED F12OL AND F12OY SEMISYNTHETIC RIBONUCLEASES | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Doscher, M.S, Glinn, M.A, Martin, P.D, Ram, M.L, Edwards, B.F.P. | | Deposit date: | 1993-08-03 | | Release date: | 1994-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of catalytically modified F120L and F120Y semisynthetic ribonucleases.
Protein Sci., 3, 1994
|
|
1WX2
 
 | | Crystal Structure of the oxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase complexed with a caddie protein prepared by the addition of hydrogenperoxide | | Descriptor: | COPPER (II) ION, MelC, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
1WY2
 
 | |
1WDN
 
 | | GLUTAMINE-BINDING PROTEIN | | Descriptor: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | Authors: | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
1SSC
 
 | | THE 1.6 ANGSTROMS STRUCTURE OF A SEMISYNTHETIC RIBONUCLEASE CRYSTALLIZED FROM AQUEOUS ETHANOL. COMPARISON WITH CRYSTALS FROM SALT SOLUTIONS AND WITH RNASE A FROM AQUEOUS ALCOHOL SOLUTIONS | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE A | | Authors: | De Mel, V.S.J, Doscher, M.S, Martin, P.D, Rodier, F, Edwards, B.F.P. | | Deposit date: | 1994-10-05 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.6 A structure of semisynthetic ribonuclease crystallized from aqueous ethanol. Comparison with crystals from salt solutions and with ribonuclease A from aqueous alcohol solutions.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1STB
 
 | | ACCOMMODATION OF INSERTION MUTATIONS ON THE SURFACE AND IN THE INTERIOR OF STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Quirk, S, Gittis, A, Keefe, L.J, Lattman, E.E. | | Deposit date: | 1994-01-17 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accommodation of insertion mutations on the surface and in the interior of staphylococcal nuclease.
Protein Sci., 3, 1994
|
|
1WEJ
 
 | | IGG1 FAB FRAGMENT (OF E8 ANTIBODY) COMPLEXED WITH HORSE CYTOCHROME C AT 1.8 A RESOLUTION | | Descriptor: | CYTOCHROME C, E8 ANTIBODY, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mylvaganam, S.E, Paterson, Y, Getzoff, E.D. | | Deposit date: | 1998-03-26 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of FabE8-cytochrome c complex to 1.8 A resolution and FabE8 to 2.26 A resolution.
J.Mol.Biol., 281, 1998
|
|
1SSP
 
 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
1SV0
 
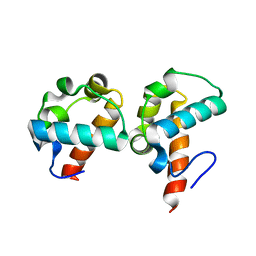 | | Crystal Structure Of Yan-SAM/Mae-SAM Complex | | Descriptor: | Ets DNA-binding protein pokkuri, modulator of the activity of Ets CG15085-PA | | Authors: | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
1X2W
 
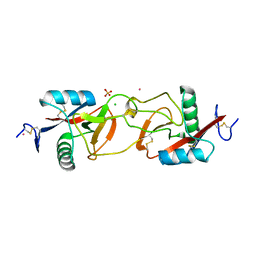 | | Crystal Structure of Apo-Habu IX-bp at pH 4.6 | | Descriptor: | CHLORIDE ION, Coagulation factor IX/X-binding protein A chain, Coagulation factor IX/factor X-binding protein B chain, ... | | Authors: | Suzuki, N, Fujimoto, Z, Morita, T, Fukamizu, A, Mizuno, H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | pH-Dependent Structural Changes at Ca(2+)-binding sites of Coagulation Factor IX-binding Protein
J.Mol.Biol., 353, 2005
|
|
1WK2
 
 | |
1QWD
 
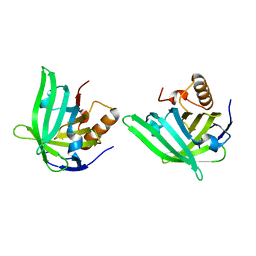 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | Deposit date: | 2003-09-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
1T2K
 
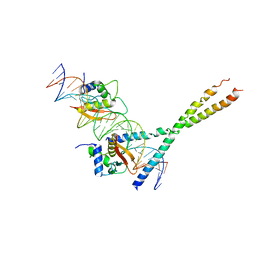 | | Structure Of The DNA Binding Domains Of IRF3, ATF-2 and Jun Bound To DNA | | Descriptor: | 31-MER, Cyclic-AMP-dependent transcription factor ATF-2, Interferon regulatory factor 3, ... | | Authors: | Panne, D, Maniatis, T, Harrison, S.C. | | Deposit date: | 2004-04-21 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer.
Embo J., 23, 2004
|
|
1X3O
 
 | |
2D4R
 
 | | Crystal structure of TTHA0849 from Thermus thermophilus HB8 | | Descriptor: | SULFATE ION, hypothetical protein TTHA0849 | | Authors: | Nakabayashi, M, Shibata, N, Kuramitsu, S, Higuchi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a conserved hypothetical protein, TTHA0849 from Thermus thermophilus HB8, at 2.4 A resolution: a putative member of the StAR-related lipid-transfer (START) domain superfamily.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1X46
 
 | | Crystal structure of a hemoglobin component (TA-VII) from Tokunagayusurika akamusi | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin component VII | | Authors: | Kuwada, T, Hasegawa, T, Sato, S, Sato, I, Ishikawa, K, Takagi, T, Shishikura, F. | | Deposit date: | 2005-05-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of two hemoglobin components from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera).
Gene, 398, 2007
|
|
1SRS
 
 | | SERUM RESPONSE FACTOR (SRF) CORE COMPLEXED WITH SPECIFIC SRE DNA | | Descriptor: | DNA (5'-D(*CP*CP*(5IU)P*TP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*CP*AP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*TP*GP*GP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*A P*AP*G)-3'), PROTEIN (SERUM RESPONSE FACTOR (SRF)) | | Authors: | Pellegrini, L, Tan, S, Richmond, T.J. | | Deposit date: | 1995-07-28 | | Release date: | 1995-07-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of serum response factor core bound to DNA.
Nature, 376, 1995
|
|
2D2N
 
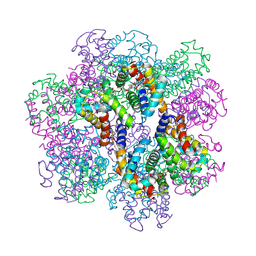 | | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi | | Descriptor: | Giant hemoglobin, A1(b) globin chain, A2(a5) globin chain, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1WNS
 
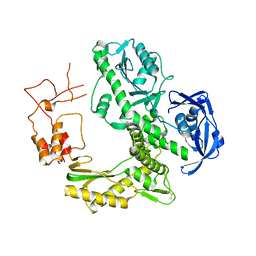 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
1SV4
 
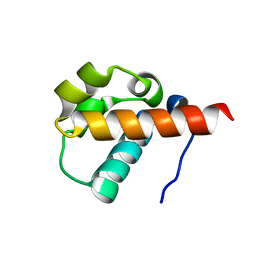 | | Crystal Structure of Yan-SAM | | Descriptor: | Ets DNA-binding protein pokkuri | | Authors: | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | Deposit date: | 2004-03-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
1SVF
 
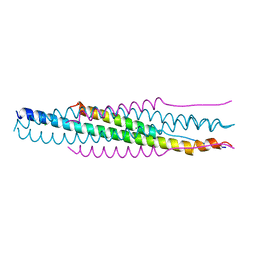 | | PARAMYXOVIRUS SV5 FUSION PROTEIN CORE | | Descriptor: | CHLORIDE ION, PROTEIN (FUSION GLYCOPROTEIN) | | Authors: | Baker, K.A, Dutch, R.E, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 1999-02-27 | | Release date: | 1999-03-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for paramyxovirus-mediated membrane fusion.
Mol.Cell, 3, 1999
|
|
2D58
 
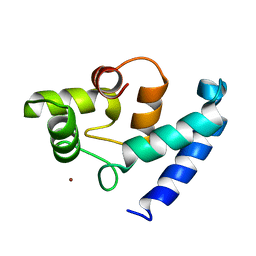 | | Human microglia-specific protein Iba1 | | Descriptor: | Allograft inflammatory factor 1, NICKEL (II) ION | | Authors: | Kamitori, S. | | Deposit date: | 2005-10-31 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structures of the Microglia/Macrophage-specific Protein Iba1 from Human and Mouse Demonstrate Novel Molecular Conformation Change Induced by Calcium binding
J.Mol.Biol., 364, 2006
|
|
1WC0
 
 | | Soluble adenylyl cyclase CyaC from S. platensis in complex with alpha, beta-methylene-ATP | | Descriptor: | ADENYLATE CYCLASE, CALCIUM ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Steegborn, C, Litvin, T.N, Levin, L.R, Buck, J, Wu, H. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bicarbonate Activation of Adenylyl Cyclase Via Promotion of Catalytic Active Site Closure and Metal Recruitment
Nat.Struct.Mol.Biol., 12, 2005
|
|
1T77
 
 | | Crystal structure of the PH-BEACH domains of human LRBA/BGL | | Descriptor: | Lipopolysaccharide-responsive and beige-like anchor protein | | Authors: | Gebauer, D, Li, J, Jogl, G, Shen, Y, Myszka, D.G, Tong, L. | | Deposit date: | 2004-05-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the PH-BEACH Domains of Human LRBA/BGL
Biochemistry, 43, 2004
|
|
1X11
 
 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
