7YP9
 
 | |
7Z03
 
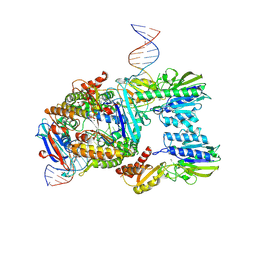 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (39-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7YZO
 
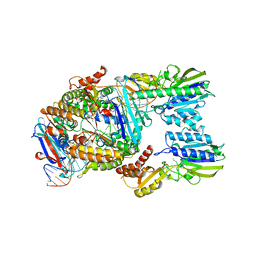 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7PLQ
 
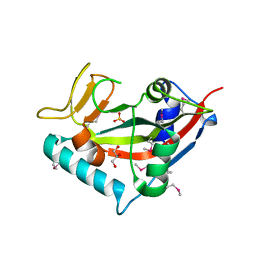 | | Crystal structure of the PARP domain of wheat SRO1 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Wirthmueller, L, Loll, B. | | Deposit date: | 2021-09-01 | | Release date: | 2021-10-27 | | Last modified: | 2022-11-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The superior salinity tolerance of bread wheat cultivar Shanrong No. 3 is unlikely to be caused by elevated Ta-sro1 poly-(ADP-ribose) polymerase activity.
Plant Cell, 34, 2022
|
|
6D6K
 
 | |
7Q2U
 
 | | The crystal structure of the HINT1 Q62A mutant. | | Descriptor: | CACODYLATE ION, HEXAETHYLENE GLYCOL, Histidine triad nucleotide-binding protein 1, ... | | Authors: | Dolot, R.M, Strom, A.M, Wagner, C.R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dynamic Long-Range Interactions Influence Substrate Binding and Catalysis by Human Histidine Triad Nucleotide-Binding Proteins (HINTs), Key Regulators of Multiple Cellular Processes and Activators of Antiviral ProTides.
Biochemistry, 61, 2022
|
|
7VCI
 
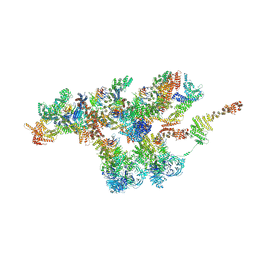 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
6D9C
 
 | |
6DOG
 
 | |
6DOU
 
 | |
6DP7
 
 | |
6DPN
 
 | |
6DAS
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | N-[(1R)-6-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)-2,3-dihydro-1H-inden-1-yl]-3-methoxy-4-methylbenzamide, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6DQA
 
 | | Linked KDM5A JMJ Domain Bound to Inhibitor N70 i.e.[2-((3-aminophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-(3-aminophenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
7VBV
 
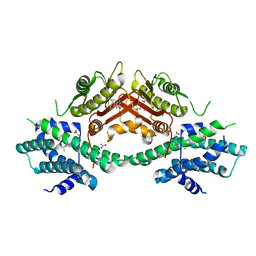 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 7 | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Fujishima, A, Adachi, T. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-based drug design of novel and highly potent pyruvate dehydrogenase kinase inhibitors.
Bioorg.Med.Chem., 52, 2021
|
|
6DEF
 
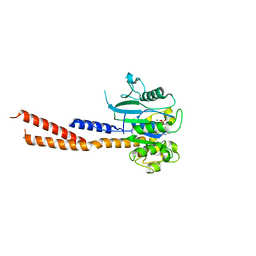 | | Vps1 GTPase-BSE fusion complexed with GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Vps1 GTPase-BSE | | Authors: | Ford, M.G.J, Varlakhanova, N.V, Brady, T.M, Chappie, J.S, Hosford, C.J. | | Deposit date: | 2018-05-11 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DID
 
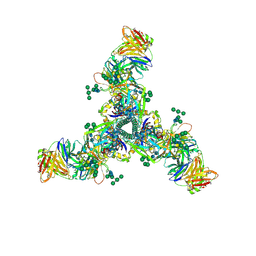 | | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Turner, H.L, Cottrell, C.A, Oyen, D, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
7PRP
 
 | |
7PFK
 
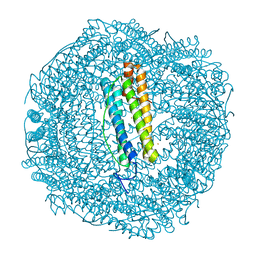 | |
6DM0
 
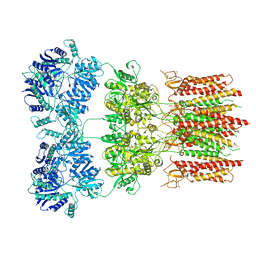 | | Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
7PFH
 
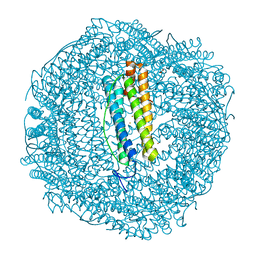 | | 2 minute Fe2+ soak structure of SynFtn E141D | | Descriptor: | ACETATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
7PTY
 
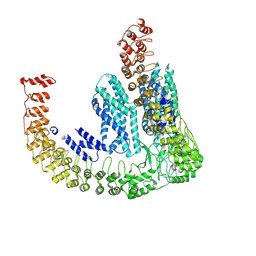 | | Delta-latroinsectotoxin dimer | | Descriptor: | Delta-latroinsectotoxin-Lt1a | | Authors: | Chen, M, Gatsogiannis, C. | | Deposit date: | 2021-09-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Molecular architecture of black widow spider neurotoxins.
Nat Commun, 12, 2021
|
|
6DPD
 
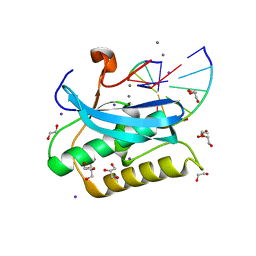 | |
7PF9
 
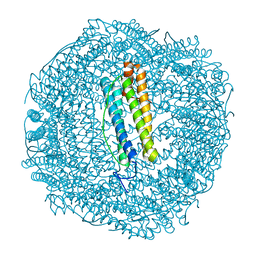 | | SynFtn Variant E141D | | Descriptor: | CHLORIDE ION, Ferritin, SODIUM ION | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
6DQC
 
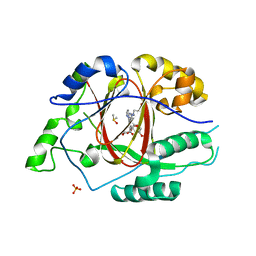 | |
