3UBY
 
 | | Crystal structure of human alklyadenine DNA glycosylase in a lower and higher-affinity complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDC)P*TP*TP*GP*CP*CP*T)-3'), DNA-3-methyladenine glycosylase | | Authors: | Setser, J.W, Lingaraju, G.M, Davis, C.A, Samson, L.D, Drennan, C.L. | | Deposit date: | 2011-10-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Searching for DNA lesions: structural evidence for lower- and higher-affinity DNA binding conformations of human alkyladenine DNA glycosylase.
Biochemistry, 51, 2012
|
|
8PSH
 
 | | HIGH RESOLUTION NMR STRUCTURE OF THE STEREOREGULAR (ALL-RP)-PHOSPHOROTHIOATE-DNA/RNA HYBRID D (G*PS*C*PS*G*PS*T*PS*C*PS*A*PS*G*PS*G)R(CCUGACGC), MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*DGP*(SC)P*(GS)P*(PST)P*(SC)P*(AS)P*(GS)P*(GS))-3'), RNA (5'-R(*CP*CP*UP*GP*AP*CP*GP*C)-3') | | Authors: | Bachelin, M, Hessler, G, Kurz, G, Hacia, J.G, Dervan, P.B, Kessler, H. | | Deposit date: | 1997-10-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Nat.Struct.Biol., 5, 1998
|
|
4ODA
 
 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
6KCS
 
 | | Crystal structure of HIRAN domain of HLTF in complex with duplex DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*AP*CP*GP*TP*AP*CP*AP*GP*T)-3'), Helicase-like transcription factor | | Authors: | Hishiki, A, Hashimoto, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of HIRAN domain of human HLTF bound to duplex DNA provides structural basis for DNA unwinding to initiate replication fork regression.
J.Biochem., 167, 2020
|
|
330D
 
 | |
7FEF
 
 | | Crystal structure of AtMBD6 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | Authors: | Wu, Z.B, Liu, K, Min, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
370D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3'), MAGNESIUM ION | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
5IFF
 
 | | Crystal structure of R.PabI-nonspecific DNA complex | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*AP*GP*TP*TP*CP*GP*AP*AP*CP*TP*AP*GP*TP*GP*C)-3'), Uncharacterized protein | | Authors: | Wang, D, Miyazono, K, Tanokura, M. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric structure of the restriction DNA glycosylase R.PabI in complex with nonspecific double-stranded DNA.
Sci Rep, 6, 2016
|
|
371D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
1DC0
 
 | |
369D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
372D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
3VK7
 
 | | Crystal structure of DNA-glycosylase bound to DNA containing 5-Hydroxyuracil | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*GP*AP*CP*GP*TP*GP*GP*AP*CP*G)-3'), FORMIC ACID, ... | | Authors: | Imamura, K, Averill, A, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-11-10 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA
J.Biol.Chem., 287, 2012
|
|
4O3N
 
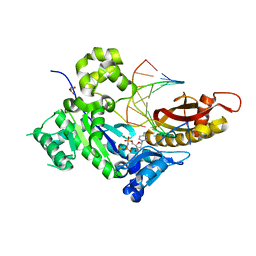 | | Crystal structure of human dna polymerase eta in ternary complex with native dna and incoming nucleotide (dcp) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
7NBP
 
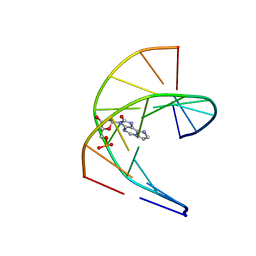 | | Solution structure of DNA duplex containing a 7,8-dihydro-8-oxo-1,N6-ethenoadenine base modification that induces exclusively A->T transversions in Escherichia coli | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*(EDA)P*CP*AP*TP*G)-3') | | Authors: | Aralov, A.V, Gubina, N, Cabrero, C, Tsvetkov, V.B, Turaev, A.V, Fedeles, B.I, Croy, R.G, Isaakova, E.A, Melnik, D, Dukova, S, Khrulev, A.A, Varizhuk, A.M, Gonzalez, C, Zatsepin, T.S, Essigmann, J.M. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 7,8-Dihydro-8-oxo-1,N6-ethenoadenine: an exclusively Hoogsteen-paired thymine mimic in DNA that induces A→T transversions in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
6XDY
 
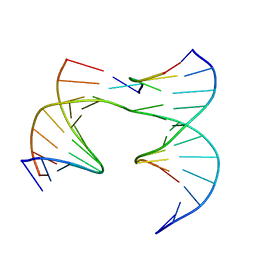 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J7 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*AP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XGO
 
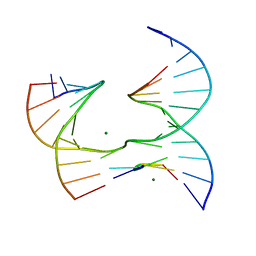 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J34 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*TP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XO8
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J31 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.195 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFF
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J22 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*GP*CP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.122 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFX
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J26 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*AP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-16 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XGN
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J33 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*AP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*GP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XEK
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J14 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6J4R
 
 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6BEK
 
 | |
8SH1
 
 | |
