1HD1
 
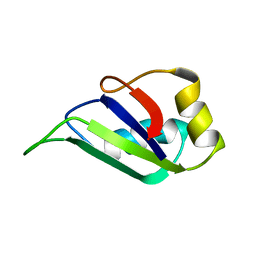 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
3V4M
 
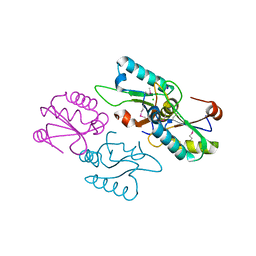 | |
6NX5
 
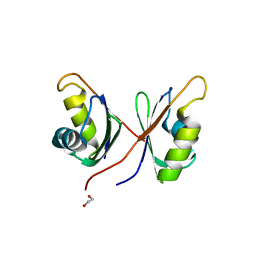 | |
6N3E
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to U2AF ligand motif 4 | | Descriptor: | FORMIC ACID, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
1P1T
 
 | |
6N3F
 
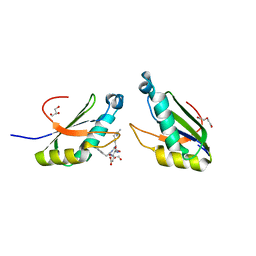 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to SF3b1 ULM5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Leach, J.R, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
3CUL
 
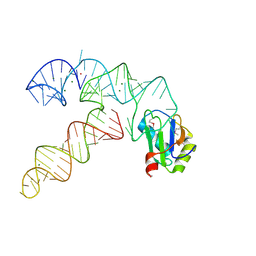 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (92-MER), ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
1HA1
 
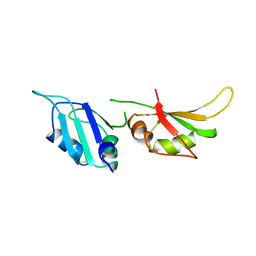 | | HNRNP A1 (RBD1,2) FROM HOMO SAPIENS | | Descriptor: | HNRNP A1 | | Authors: | Shamoo, Y, Krueger, U, Rice, L, Williams, K.R, Steitz, T.A. | | Deposit date: | 1996-10-30 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
3D2W
 
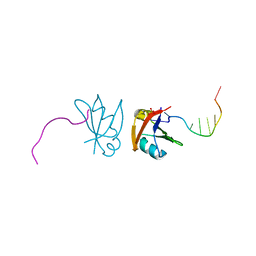 | | Crystal structure of mouse TDP-43 RRM2 domain in complex with DNA | | Descriptor: | DNA (5'-D(*DGP*DTP*DTP*DGP*DAP*DGP*DCP*DGP*DTP*DT)-3'), PHOSPHATE ION, TAR DNA-binding protein 43 | | Authors: | Kuo, P.H, Yuan, H.S. | | Deposit date: | 2008-05-09 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into TDP-43 in nucleic-acid binding and domain interactions
Nucleic Acids Res., 37, 2009
|
|
3CUN
 
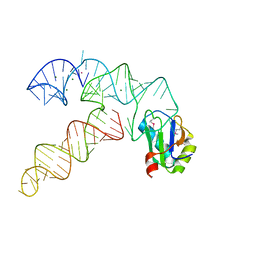 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
1NO8
 
 | | SOLUTION STRUCTURE OF THE NUCLEAR FACTOR ALY RBD DOMAIN | | Descriptor: | ALY | | Authors: | Perez-Alvarado, G.C, Martinez-Yamout, M, Allen, M.M, Grosschedl, R, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-01-15 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Nuclear Factor ALY: Insights into Post-Transcriptional
Regulatory and mRNA Nuclear Export Processes
Biochemistry, 42, 2003
|
|
1NU4
 
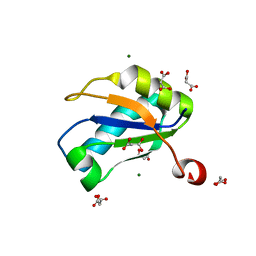 | | U1A RNA binding domain at 1.8 angstrom resolution reveals a pre-organized C-terminal helix | | Descriptor: | MAGNESIUM ION, MALONIC ACID, U1A RNA binding domain | | Authors: | Rupert, P.B, Xiao, H, Ferre-D'Amare, A.R. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | U1A RNA-binding domain at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3G8S
 
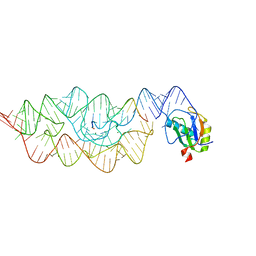 | | Crystal structure of the pre-cleaved Bacillus anthracis glmS ribozyme | | Descriptor: | GLMS RIBOZYME, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
1OIA
 
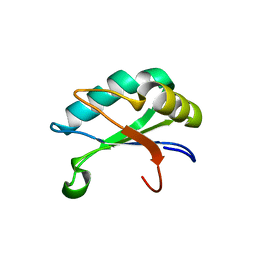 | | U1A rnp domain 1-95 | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Nagai, K, Evans, P.R. | | Deposit date: | 2003-06-12 | | Release date: | 2003-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the RNA-Binding Domain of the U1 Small Nuclear Ribonucleoprotein A
Nature, 348, 1990
|
|
3G8T
 
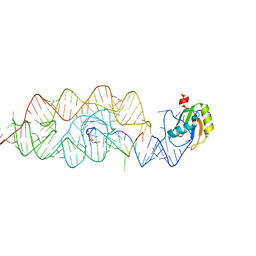 | | Crystal structure of the G33A mutant Bacillus anthracis glmS ribozyme bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
1PO6
 
 | |
3VAG
 
 | | Structure of U2AF65 variant with BrU3C2 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*U*CP*(BRU)P*UP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAH
 
 | | Structure of U2AF65 variant with BrU3C4 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(P*UP*UP*(BRU)P*CP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3G9C
 
 | | Crystal structure of the product Bacillus anthracis glmS ribozyme | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-13 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | Descriptor: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | Authors: | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
3G96
 
 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
5DET
 
 | | X-ray structure of human RBPMS in complex with the RNA | | Descriptor: | RNA (5'-R(*UP*CP*AP*C)-3'), RNA (5'-R(P*UP*CP*AP*CP*U)-3'), RNA-binding protein with multiple splicing, ... | | Authors: | Teplova, M, Farazi, T.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
5KW1
 
 | | Crystal Structure of the Two Tandem RRM Domains of PUF60 Bound to a Modified AdML Pre-mRNA 3' Splice Site Analogue | | Descriptor: | CHLORIDE ION, DNA/RNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5D77
 
 | | Structure of Mip6 RRM3 Domain | | Descriptor: | CITRIC ACID, NITRATE ION, RNA-binding protein MIP6, ... | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mip6 RRM3 domain at 1.3
To Be Published
|
|
5KVY
 
 | | CRYSTAL STRUCTURE OF THE TWO TANDEM RRM DOMAINS OF PUF60 BOUND TO A PORTION OF AN ADML PRE-MRNA 3' SPLICE SITE ANALOG | | Descriptor: | CHLORIDE ION, DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Hsiao, H.-H, Crichlow, G.V, Albright, R.A, Murphy, J.W, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
