4QL1
 
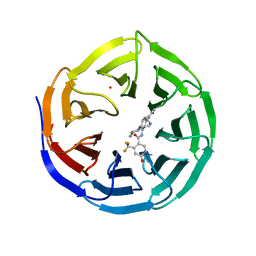 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
1WEU
 
 | | Solution structure of PHD domain in ING1-like protein BAC25009 | | Descriptor: | ZINC ION, inhibitor of growth family, member 4 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in ING1-like protein BAC25009
To be Published
|
|
7VVZ
 
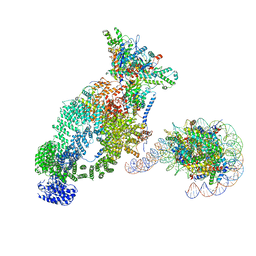 | | NuA4 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Qu, K, Chen, Z. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
7VVU
 
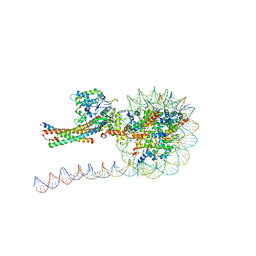 | | NuA4 HAT module bound to the nucleosome | | Descriptor: | CARBOXYMETHYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z, Qu, K. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
1WE9
 
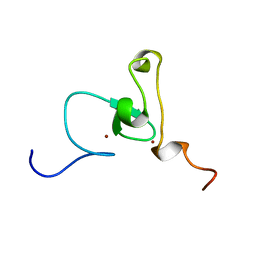 | | Solution structure of PHD domain in nucleic acid binding protein-like NP_197993 | | Descriptor: | PHD finger family protein, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in nucleic acid binding protein-like NP_197993
To be Published
|
|
1WES
 
 | | Solution structure of PHD domain in inhibitor of growth family, member 1-like | | Descriptor: | ZINC ION, inhibitor of growth family, member 1-like | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in inhibitor of growth family, member 1-like
To be Published
|
|
6OEM
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | Descriptor: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6I8L
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
1X4I
 
 | | Solution structure of PHD domain in inhibitor of growth protein 3 (ING3) | | Descriptor: | Inhibitor of growth protein 3, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in inhibitor of growth protein 3 (ING3)
To be Published
|
|
4RIQ
 
 | |
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OET
 
 | | Cryo-EM structure of mouse RAG1/2 STC complex | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (39-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEP
 
 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7XE1
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE3
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, CITRATE ANION, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE2
 
 | | Crystal structure of LSD2 in complex with trans-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
6I8Y
 
 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6OES
 
 | | Cryo-EM structure of mouse RAG1/2 STC complex (without NBD domain) | | Descriptor: | CALCIUM ION, DNA (34-MER), DNA (35-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6I8B
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6L87
 
 | |
9GII
 
 | | Jumonji domain-containing protein 2A with crystallization epitope mutation R913A | | Descriptor: | GLYCEROL, Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
9GD2
 
 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD1
 
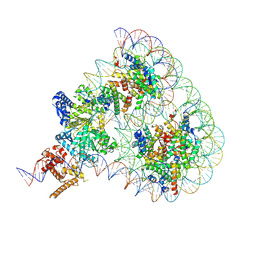 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD3
 
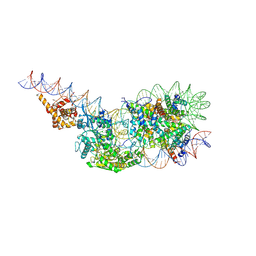 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GP4
 
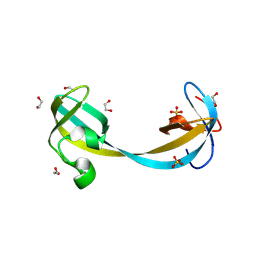 | | Jumonji domain-containing protein 2A with crystallization epitope mutations Q953E:A958D | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
