7L7E
 
 | |
6T4D
 
 | | Crystal structure of Plasmodium falciparum Morn1 | | Descriptor: | Morn1, ZINC ION | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
7L7D
 
 | |
5MYB
 
 | | Homodimerization of Tie2 Fibronectin-like domains 2 and 3 in space group P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiopoietin-1 receptor, ... | | Authors: | Leppanen, V.-M, Saharinen, P, Alitalo, K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7BB7
 
 | | AVP-V2R-Galphas-beta1-gamma2-Nb35(T state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Bous, J, Mouillac, B, Bron, P, Granier, S, Floquet, N, Leyrat, C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-electron microscopy structure of the antidiuretic hormone arginine-vasopressin V2 receptor signaling complex.
Sci Adv, 7, 2021
|
|
1AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 10 STRUCTURES | | Descriptor: | PHENOL, R6 INSULIN HEXAMER, ZINC ION | | Authors: | Chang, X, Jorgensen, A.M.M, Bardrum, P, Led, J.J. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the R6 human insulin hexamer.
Biochemistry, 36, 1997
|
|
6VCB
 
 | | Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator | | Descriptor: | 1-[(1R)-1-(2,6-dichloro-3-methoxyphenyl)ethyl]-6-{2-[(2R)-piperidin-2-yl]phenyl}-1H-benzimidazole, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Feng, D, Bueno, A, Kobilka, B, Sloop, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into probe-dependent positive allosterism of the GLP-1 receptor.
Nat.Chem.Biol., 16, 2020
|
|
6TO9
 
 | |
7BB6
 
 | | AVP-V2R-Galphas-beta1-gamma2-Nb35 (L state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Bous, J, Mouillac, B, Bron, P, Granier, S, Floquet, N, Leyrat, C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structure of the antidiuretic hormone arginine-vasopressin V2 receptor signaling complex.
Sci Adv, 7, 2021
|
|
6MN6
 
 | |
7BJL
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-142 | | Descriptor: | 14-3-3 protein sigma, 3-[(3~{S})-3-methoxypiperidin-1-yl]-4-nitro-benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJW
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-154 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-(4-oxidanylpiperidin-1-yl)benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJF
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-069 | | Descriptor: | (5-methyl-2-nitro-phenyl) cyclobutanecarboxylate, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BIQ
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-180 | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{R})-2-methyl-3,4-dihydro-2~{H}-quinolin-1-yl]sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
5MEB
 
 | | Crystal structure of yeast Cdt1 C-terminal domain | | Descriptor: | Cell division cycle protein CDT1, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
6CKN
 
 | | Crystal structure of an AF10 fragment | | Descriptor: | Protein AF-10, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and functional analysis of the DOT1L-AF10 complex reveals mechanistic insights into MLL-AF10-associated leukemogenesis.
Genes Dev., 32, 2018
|
|
7BKH
 
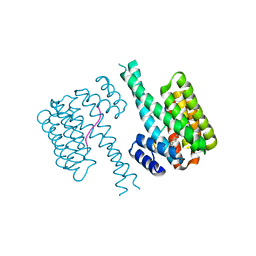 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-153 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-[(3S)-3-oxidanylpiperidin-1-yl]benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
5K3H
 
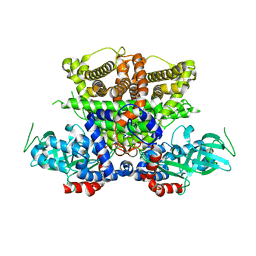 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-II | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7BI3
 
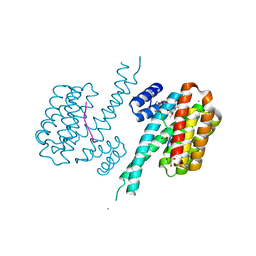 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-179 | | Descriptor: | 14-3-3 protein sigma, 4-[(6-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
5MM0
 
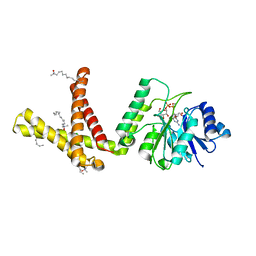 | | Dolichyl phosphate mannose synthase in complex with GDP-mannose and Mn2+ (donor complex) | | Descriptor: | CHLORIDE ION, Dolichol monophosphate mannose synthase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Gandini, R, Reichenbach, T, Tan, T.C, Divne, C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for dolichylphosphate mannose biosynthesis.
Nat Commun, 8, 2017
|
|
7BIY
 
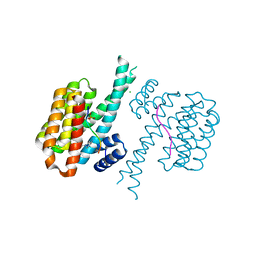 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-175 | | Descriptor: | 1-(4-methanoylphenyl)carbonylpiperidine-4-carbonitrile, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
6JT9
 
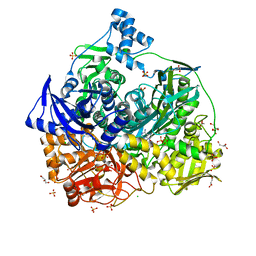 | | Crystal Structure of D464A mutant of FGAM Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6CRK
 
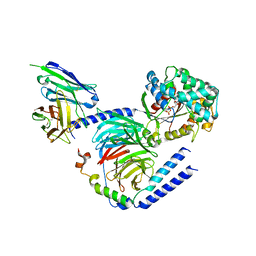 | | Heterotrimeric G-protein in complex with an antibody fragment | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Maeda, S, Dawson, R, Kobilka, B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an antibody fragment that stabilizes GPCR/G-protein complexes.
Nat Commun, 9, 2018
|
|
6CQK
 
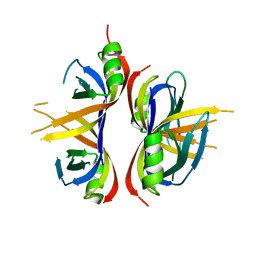 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae, Rim1 (Form1) | | Descriptor: | SsDNA-binding protein essential for mitochondrial genome maintenance | | Authors: | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
5K5T
 
 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
