2GQS
 
 | | SAICAR Synthetase Complexed with CAIR-Mg2+ and ADP | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, ... | | Authors: | Ginder, N.D, Honzatko, R.B. | | Deposit date: | 2006-04-21 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Nucleotide Complexes of Escherichia coli Phosphoribosylaminoimidazole Succinocarboxamide Synthetase.
J.Biol.Chem., 281, 2006
|
|
2PD5
 
 | | Human aldose reductase mutant V47I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDC
 
 | | Human aldose reductase mutant F121P complexed with IDD393. | | Descriptor: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDF
 
 | | Human aldose reductase mutant L300P complexed with zopolrestat. | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDL
 
 | |
2PDX
 
 | | Human aldose reductase double mutant S302R-C303D complexed with zopolrestat. | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDP
 
 | | Human aldose reductase mutant S302R complexed with IDD 393. | | Descriptor: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDK
 
 | | Human aldose reductase mutant L301M complexed with sorbinil. | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SORBINIL | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDG
 
 | | Human aldose reductase with uracil-type inhibitor at 1.42A. | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {3-[(5-CHLORO-1,3-BENZOTHIAZOL-2-YL)METHYL]-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL}ACETIC ACID | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
4V1L
 
 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1S3P
 
 | |
4V44
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
3UDJ
 
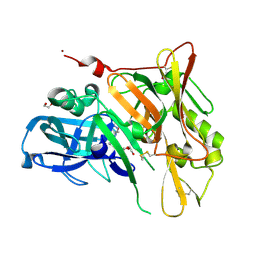 | | Crystal Structure of BACE with Compound 5 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3CKU
 
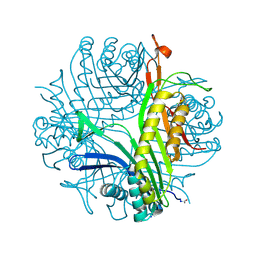 | |
4G4P
 
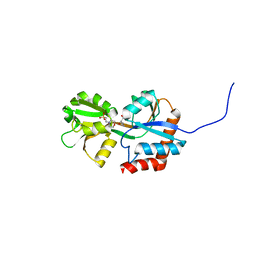 | | Crystal structure of glutamine-binding protein from Enterococcus faecalis at 1.5 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Amino acid ABC transporter, amino acid-binding/permease protein, ... | | Authors: | Fulyani, F, Guskov, A, Zagar, A.V, Slotboom, D.-J, Poolman, B. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
6ZHC
 
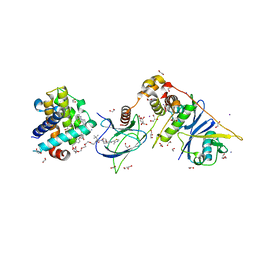 | | PROTAC6 mediated complex of VHL:EloB:EloC and Bcl-xL | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]-5-[3-[4-[3-[2-[2-[2-[2-[2-[3-[[(2~{S})-3,3-dimethyl-1-[(2~{S},4~{R})-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-3-oxidanylidene-propoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]prop-1-ynyl]phenoxy]propyl]-1,3-thiazole-4-carboxylic acid, Bcl-2-like protein 1, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into PROTAC-Mediated Degradation of Bcl-xL.
Acs Chem.Biol., 15, 2020
|
|
3CXO
 
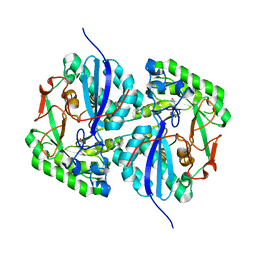 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and 3-deoxy-L-rhamnonate | | Descriptor: | (2R,4S)-2,4,7-trihydroxyheptanoic acid, 3,6-dideoxy-L-arabino-hexonic acid, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Hubbard, B.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-24 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
6WCV
 
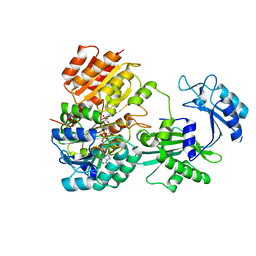 | | Tartryl-CoA bound to human GTP-specific succinyl-CoA synthetase | | Descriptor: | (3S,5S,9R,20R,21R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,20, 21-pentahydroxy-8,8-dimethyl-10,14,19-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphadocosan-22-oic acid 3,5-dioxide (non-preferred name), Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tartryl-CoA inhibits succinyl-CoA synthetase
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7T4Z
 
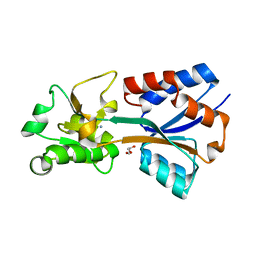 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in ligand-free form | | Descriptor: | AMMONIUM ION, GLYCEROL, Molybdate-binding periplasmic protein, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
1S78
 
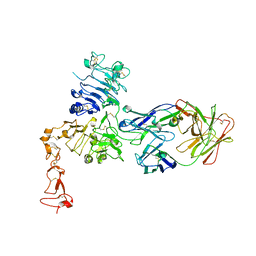 | | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, ... | | Authors: | Franklin, M.C, Carey, K.D, Vajdos, F.F, Leahy, D.J, de Vos, A.M, Sliwkowski, M.X. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex.
Cancer Cell, 5, 2004
|
|
4WGI
 
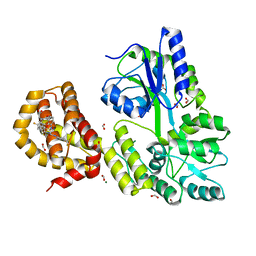 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
4WH7
 
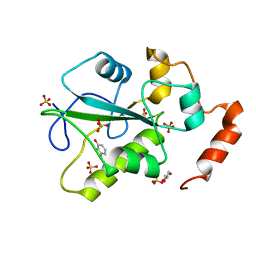 | | Structure of the CDC25B Phosphatase Catalytic Domain with Bound Ligand | | Descriptor: | 2-fluoro-4-hydroxybenzonitrile, GLYCEROL, M-phase inducer phosphatase 2, ... | | Authors: | Lund, G.L, Dudkin, S, Borkin, D, Ni, W, Grembecka, J, Cierpicki, T. | | Deposit date: | 2014-09-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction.
Acs Chem.Biol., 10, 2015
|
|
7ALJ
 
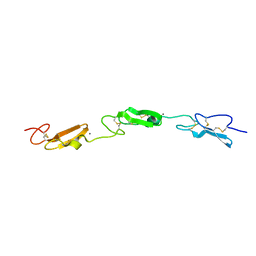 | | Structure of Drosophila Notch EGF domains 11-13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus Notch protein, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
7ALK
 
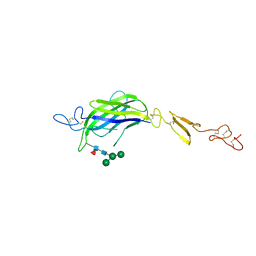 | | Structure of Drosophila C2-DSL-EGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neurogenic locus protein delta, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
7ALT
 
 | | Structure of Drosophila Serrate C2-DSL-EGF1-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
