2BAM
 
 | |
2B1C
 
 | |
2B1K
 
 | | Crystal structure of E. coli CcmG protein | | Descriptor: | Thiol:disulfide interchange protein dsbE | | Authors: | Ouyang, N, Gao, Y.G, Hu, H.Y, Xia, Z.X. | | Deposit date: | 2005-09-15 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli CcmG and its mutants reveal key roles of the N-terminal beta-sheet and the fingerprint region
Proteins, 65, 2006
|
|
2AHD
 
 | |
2FBV
 
 | | WRN exonuclease, Mn complex | | Descriptor: | MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2AS0
 
 | | Crystal Structure of PH1915 (APC 5817): A Hypothetical RNA Methyltransferase | | Descriptor: | hypothetical protein PH1915 | | Authors: | Sun, W, Xu, X, Pavlova, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
Protein Sci., 14, 2005
|
|
2FCP
 
 | | FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) FROM E.COLI | | Descriptor: | 2-TRIDECANOYLOXY-PENTADECANOIC ACID, 3-OXO-PENTADECANOIC ACID, ACETOACETIC ACID, ... | | Authors: | Hofmann, E, Ferguson, A.D, Diederichs, K, Welte, W. | | Deposit date: | 1998-10-15 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide.
Science, 282, 1998
|
|
2FBT
 
 | | WRN exonuclease | | Descriptor: | ACETIC ACID, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2COL
 
 | | Crystal structure analysis of CyaA/C-Cam with Pyrophosphate | | Descriptor: | Bifunctional hemolysin-adenylate cyclase, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Tang, W.J, Shen, Y. | | Deposit date: | 2005-05-18 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin
Embo J., 24, 2005
|
|
2CUZ
 
 | |
2CY2
 
 | | Crystal structure of TTHA1209 in complex with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, probable acetyltransferase | | Authors: | Kaminishi, T, Takemoto, C, Uchikubo-Kamo, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TTHA1209 in complex with acetyl coenzyme A
To be Published
|
|
2FMT
 
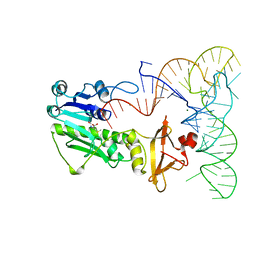 | | METHIONYL-TRNAFMET FORMYLTRANSFERASE COMPLEXED WITH FORMYL-METHIONYL-TRNAFMET | | Descriptor: | FORMYL-METHIONYL-TRNAFMET2, MAGNESIUM ION, METHIONYL-TRNA FMET FORMYLTRANSFERASE, ... | | Authors: | Schmitt, E, Mechulam, Y, Blanquet, S. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet.
EMBO J., 17, 1998
|
|
1HUT
 
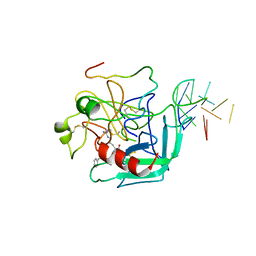 | | THE STRUCTURE OF ALPHA-THROMBIN INHIBITED BY A 15-MER SINGLE-STRANDED DNA APTAMER | | Descriptor: | ALPHA-Thrombin heavy chain, ALPHA-Thrombin light chain, D-phenylalanyl-N-[(3S)-6-carbamimidamido-1-chloro-2-oxohexan-3-yl]-L-prolinamide, ... | | Authors: | Padmanabhan, K, Padmanabhan, K.P, Ferrara, J.D, Sadler, J.E, Tulinsky, A. | | Deposit date: | 1993-05-27 | | Release date: | 1994-06-22 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer.
J.Biol.Chem., 268, 1993
|
|
1EGG
 
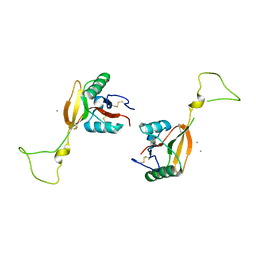 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
1HVR
 
 | | RATIONAL DESIGN OF POTENT, BIOAVAILABLE, NONPEPTIDE CYCLIC UREAS AS HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-HEXAHYDRO-5,6-DIHYDROXY-1,3-BIS[2-NAPHTHYL-METHYL]-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPIN-2-ONE | | Authors: | Chang, C.-H. | | Deposit date: | 1994-02-14 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors.
Science, 263, 1994
|
|
1EH6
 
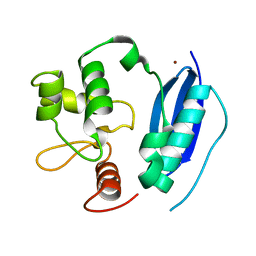 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE | | Descriptor: | O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE, ZINC ION | | Authors: | Daniels, D.S, Tainer, J.A. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding.
EMBO J., 19, 2000
|
|
1HZP
 
 | | Crystal Structure of the Myobacterium Tuberculosis Beta-Ketoacyl-Acyl Carrier Protein Synthase III | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE III, GLYCEROL, LAURIC ACID | | Authors: | Scarsdale, J.N, Kazanina, G, He, X, Reynolds, K.A, Wright, H.T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-01-25 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III
J.Biol.Chem., 276, 2001
|
|
1EHW
 
 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
1EJG
 
 | | CRAMBIN AT ULTRA-HIGH RESOLUTION: VALENCE ELECTRON DENSITY. | | Descriptor: | CRAMBIN (PRO22,SER22/LEU25,ILE25) | | Authors: | Jelsch, C, Teeter, M.M, Lamzin, V, Pichon-Lesme, V, Blessing, B, Lecomte, C. | | Deposit date: | 2000-03-02 | | Release date: | 2000-04-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (0.54 Å) | | Cite: | Accurate protein crystallography at ultra-high resolution: valence electron distribution in crambin.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1HZH
 
 | | CRYSTAL STRUCTURE OF THE INTACT HUMAN IGG B12 WITH BROAD AND POTENT ACTIVITY AGAINST PRIMARY HIV-1 ISOLATES: A TEMPLATE FOR HIV VACCINE DESIGN | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN,Uncharacterized protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Burton, D.R, Wilson, I.A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-08-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design.
Science, 293, 2001
|
|
1HZX
 
 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Teller, D.C, Okada, T, Behnke, C.A, Palczewski, K, Stenkamp, R.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs).
Biochemistry, 40, 2001
|
|
1HLP
 
 | |
1EMK
 
 | |
1EYG
 
 | |
1EOF
 
 | | CRYSTAL STRUCTURE OF THE N136A MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
