4J7B
 
 | | Crystal structure of polo-like kinase 1 | | Descriptor: | 205 kDa microtubule-associated protein, Polo-like kinase | | Authors: | Xu, J, Shen, C, Quan, J, Wang, T. | | Deposit date: | 2013-02-13 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the inhibition of Polo-like kinase 1
Nat.Struct.Mol.Biol., 20, 2013
|
|
4J9O
 
 | | Human DNA polymerase eta-DNA ternary complex: primer extension after a T:G mispair | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*AP*CP*GP*TP*CP*AP*TP*G)-3'), ... | | Authors: | Zhao, Y, Gregory, M, Biertumpfel, C, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2013-02-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JP4
 
 | | Mmp13 in complex with a reverse hydroxamate Zn-binder | | Descriptor: | CALCIUM ION, Collagenase 3, N-[(2S)-4-(5-fluoropyrimidin-2-yl)-1-({4-[5-(2,2,2-trifluoroethoxy)pyrimidin-2-yl]piperazin-1-yl}sulfonyl)butan-2-yl]-N-hydroxyformamide, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JAX
 
 | | Crystal structure of dimeric KlHxk1 in crystal form X | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Strater, N, Kettner, K, Otto, A, Lilie, H, Golbik, R.P, Kriegel, T.M. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | In vivo phosphorylation and in vitro autophosphorylation-inactivation of Kluyveromyces lactis hexokinase KlHxk1.
Biochem.Biophys.Res.Commun., 435, 2013
|
|
4JEL
 
 | | Structure of MilB Streptomyces rimofaciens CMP N-glycosidase | | Descriptor: | CMP/hydroxymethyl CMP hydrolase, SULFATE ION | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
4JM9
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-amino-1-methylpyridinium | | Descriptor: | 1-METHYL-1,6-DIHYDROPYRIDIN-3-AMINE, Cytochrome c peroxidase, IODIDE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JN0
 
 | |
4JNU
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group P21 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4J8A
 
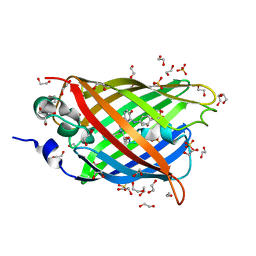 | | Irradiated-state structure of sfGFP containing the unnatural amino acid p-azido-phenylalanine at residue 145 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Reddington, S.C, Jones, D.D, Rizkallah, P.J, Tippmann, E.M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Different Photochemical Events of a Genetically Encoded Phenyl Azide Define and Modulate GFP Fluorescence.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4J9K
 
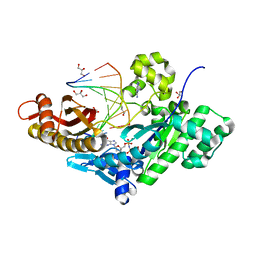 | | Human DNA polymerase eta-DNA ternary complex: misincorporation G opposite T after a T at the primer 3' end (TA/G) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA, DNA polymerase eta, ... | | Authors: | Zhao, Y, Gregory, M, Biertumpfel, C, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2013-02-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1QIJ
 
 | |
4JL5
 
 | |
4JMS
 
 | |
4JQN
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Hydroxybenzaldehyde | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, P-HYDROXYBENZALDEHYDE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JVH
 
 | | Structure of the star domain of quaking protein in complex with RNA | | Descriptor: | Protein quaking, RNA (5'-R(*UP*UP*CP*AP*CP*UP*AP*AP*CP*AP*A)-3'), SULFATE ION | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
2QI9
 
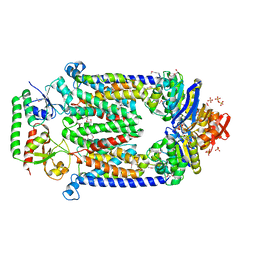 | | ABC-transporter BtuCD in complex with its periplasmic binding protein BtuF | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PHOSPHATE ION, ... | | Authors: | Hvorup, R.N, Goetz, B.A, Niederer, M, Hollenstein, K, Perozo, E, Locher, K.P. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF.
Science, 317, 2007
|
|
2R3M
 
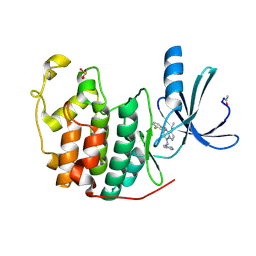 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | Cell division protein kinase 2, N-((2-aminopyrimidin-5-yl)methyl)-5-(2,6-difluorophenyl)-3-ethylpyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
1QIK
 
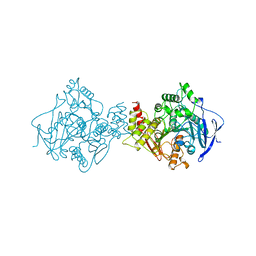 | |
4J5X
 
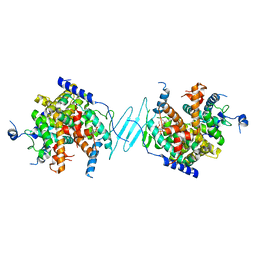 | | Crystal Structure of the SR12813-bound PXR/RXRalpha LBD Heterotetramer Complex | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1, Retinoic acid receptor RXR-alpha, ... | | Authors: | Wallace, B.D, Betts, L, Redinbo, M.R. | | Deposit date: | 2013-02-10 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Analysis of the Human Nuclear Xenobiotic Receptor PXR in Complex with RXRalpha.
J.Mol.Biol., 425, 2013
|
|
2R3F
 
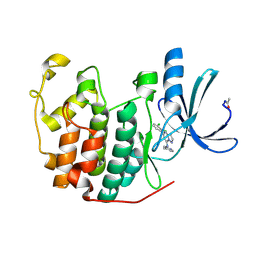 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2,3-dichlorophenyl)-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2QOR
 
 | |
2R3Q
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-((3-bromo-5-o-tolylpyrazolo[1,5-a]pyrimidin-7-ylamino)methyl)pyridine 1-oxide, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2QTY
 
 | |
2QZQ
 
 | | Crystal structure of C-terminal of Aida | | Descriptor: | Axin interactor, dorsalization associated protein | | Authors: | Zheng, L.S, Wu, J.W. | | Deposit date: | 2007-08-17 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of C-terminal of Aida
To be Published
|
|
4FUW
 
 | | Crystal structure of Ego3 mutant | | Descriptor: | Protein SLM4, SULFATE ION | | Authors: | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
