4KGO
 
 | |
2A1O
 
 | |
2A8V
 
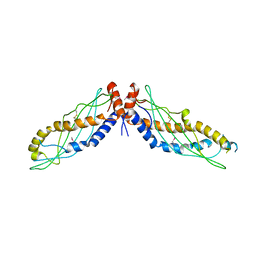
 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | Descriptor: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | Authors: | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | Deposit date: | 1998-11-08 | | Release date: | 1999-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
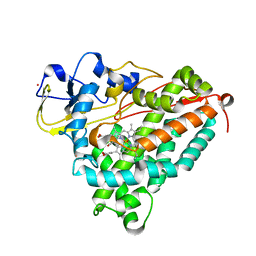
2A6Q
 
 | | Crystal structure of YefM-YoeB complex | | Descriptor: | Antitoxin yefM, Toxin yoeB | | Authors: | Kamada, K, Hanaoka, F. | | Deposit date: | 2005-07-04 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Change in the Catalytic Site of the Ribonuclease YoeB Toxin by YefM Antitoxin
Mol.Cell, 19, 2005
|
|
2ABL
 
 | |
2AFN
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE AND A COPPER SITE MUTANT, M150E, THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
2AEZ
 
 | | Crystal structure of fructan 1-exohydrolase IIa (E201Q) from Cichorium intybus in complex with 1-kestose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Lammens, W, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
1ZVM
 
 | | Crystal structure of human CD38: cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase | | Descriptor: | ADP-ribosyl cyclase 1, SULFATE ION | | Authors: | Shi, W, Yang, T, Almo, S.C, Schramm, V.L, Sauve, A. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human CD38: Cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase
To be Published
|
|
208L
 
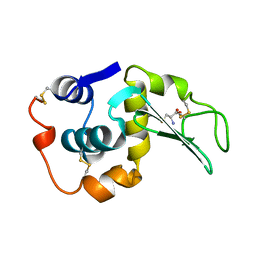 | | MUTANT HUMAN LYSOZYME C77A | | Descriptor: | CYSTEINE, LYSOZYME | | Authors: | Matsushima, M, Song, H. | | Deposit date: | 1996-03-26 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A role of PDI in the reductive cleavage of mixed disulfides.
J.Biochem.(Tokyo), 120, 1996
|
|
1ZW9
 
 | |
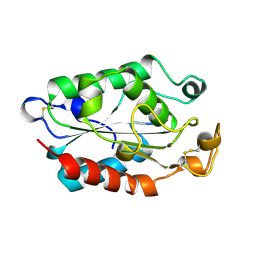
1XZD
 
 | |
1XXB
 
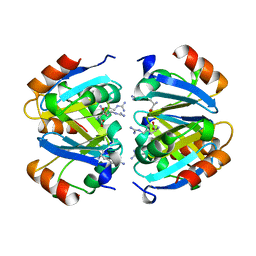 | | C-TERMINAL DOMAIN OF ESCHERICHIA COLI ARGININE REPRESSOR/ L-ARGININE COMPLEX | | Descriptor: | ARGININE, ARGININE REPRESSOR | | Authors: | Van Duyne, G.D, Ghosh, G, Maas, W.K, Sigler, P.B. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli.
J.Mol.Biol., 256, 1996
|
|
1XYC
 
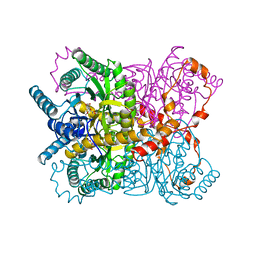 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | 3-O-METHYLFRUCTOSE IN LINEAR FORM, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XNB
 
 | |
1XZF
 
 | |
1XQK
 
 | | Effect of a Y265F Mutant on the Transamination Based Cycloserine Inactivation of Alanine Racemase | | Descriptor: | (5-HYDROXY-4-{[(3-HYDROXYISOXAZOL-4-YL)AMINO]METHYL}-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Alanine racemase | | Authors: | Fenn, T.D, Holyoak, T, Stamper, G.F, Ringe, D. | | Deposit date: | 2004-10-12 | | Release date: | 2005-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Effect of a Y265F Mutant on the Transamination-Based Cycloserine Inactivation of Alanine Racemase
Biochemistry, 44, 2005
|
|
1XRS
 
 | | Crystal structure of Lysine 5,6-Aminomutase in complex with PLP, cobalamin, and 5'-deoxyadenosine | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-lysine 5,6-aminomutase alpha subunit, ... | | Authors: | Berkovitch, F, Behshad, E, Tang, K.H, Enns, E.A, Frey, P.A, Drennan, C.L. | | Deposit date: | 2004-10-15 | | Release date: | 2004-11-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XVF
 
 | |
1XYB
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | D-glucose, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XZB
 
 | |
1XZK
 
 | |
1XKJ
 
 | | BACTERIAL LUCIFERASE BETA2 HOMODIMER | | Descriptor: | BETA2 LUCIFERASE | | Authors: | Tanner, J.J, Krause, K.L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial luciferase beta 2 homodimer: implications for flavin binding.
Biochemistry, 36, 1997
|
|
1XGN
 
 | |
1XSM
 
 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
1XWL
 
 | |
