3SG8
 
 | |
1ULG
 
 | | CGL2 in complex with Thomsen-Friedenreich antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
4U0H
 
 | | Crystal Structure of M. tuberculosis ClpP1P1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, SULFATE ION | | Authors: | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2479 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7MJF
 
 | | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site | | Descriptor: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, (4S)-4-hydroxy-2-oxoheptanedioic acid, 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J, Frampton, R.A, Board, A.J, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site
To Be Published
|
|
4DVF
 
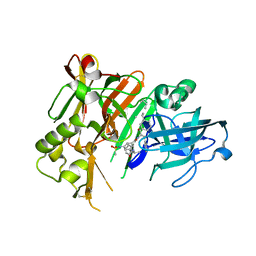 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
2Z87
 
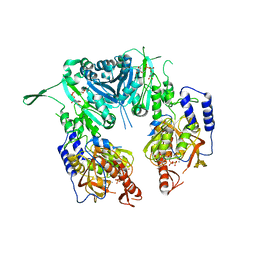 | | Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP | | Descriptor: | Chondroitin synthase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Osawa, T, Sugiura, N, Shimada, H, Hirooka, R, Tsuji, A, Kimura, M, Kimata, K, Kakuta, Y. | | Deposit date: | 2007-09-03 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chondroitin polymerase from Escherichia coli K4.
Biochem. Biophys. Res. Commun., 378, 2009
|
|
5G1Z
 
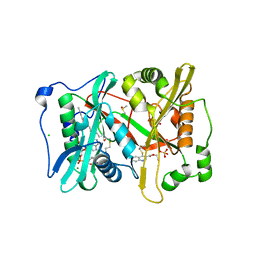 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 1) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G22
 
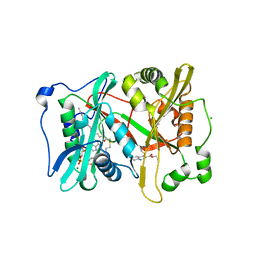 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 26) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
3LHQ
 
 | | DNA-binding transcriptional repressor AcrR from Salmonella typhimurium. | | Descriptor: | 1,2-ETHANEDIOL, AcrAB operon repressor (TetR/AcrR family), DI(HYDROXYETHYL)ETHER | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray crystal structure of DNA-binding transcriptional repressor AcrR from Salmonella typhimurium.
To be Published
|
|
3C5T
 
 | |
1CS1
 
 | |
8G3H
 
 | |
5AL4
 
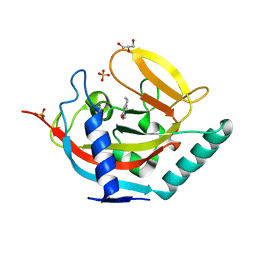 | | Crystal structure of TNKS2 in complex with 2-(4-methylpiperazin-1-yl)- 3,4,5,6,7,8-hexahydroquinazolin-4-one | | Descriptor: | 2-(4-METHYLPIPERAZIN-1-YL)-3,4,5,6,7,8-HEXAHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Lehtio, L. | | Deposit date: | 2015-03-06 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Nonplanar Tankyrase Inhibiting Nicotinamide Mimics.
Bioorg.Med.Chem., 23, 2015
|
|
1SRU
 
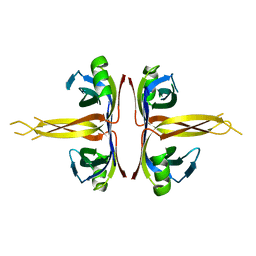 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
3C3Z
 
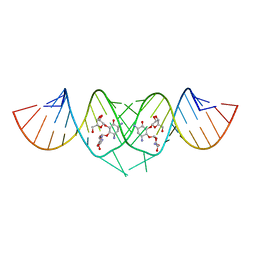 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to ribostamycin | | Descriptor: | HIV-1 subtype F genomic RNA, RIBOSTAMYCIN | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
1BQG
 
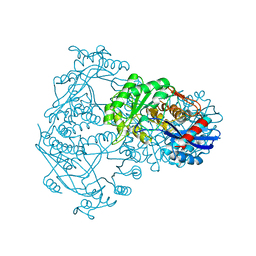 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | Descriptor: | D-GLUCARATE DEHYDRATASE | | Authors: | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 1998-08-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
3BM4
 
 | | Crystal Structure of Human ADP-ribose Pyrophosphatase NUDT5 In complex with magnesium and AMPcpr | | Descriptor: | ADP-sugar pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MAGNESIUM ION | | Authors: | Zha, M, Guo, Q, Zhang, Y, Zhong, C, Ou, Y, Ding, J. | | Deposit date: | 2007-12-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of ADP-Ribose Hydrolysis By Human NUDT5 From Structural and Kinetic Studies
J.Mol.Biol., 379, 2008
|
|
7M7G
 
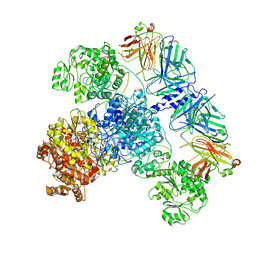 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
7M7H
 
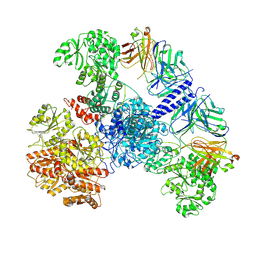 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1' | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
7M7F
 
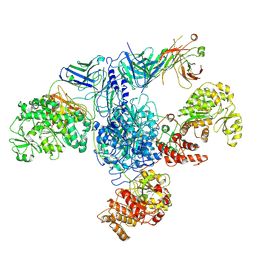 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
3C59
 
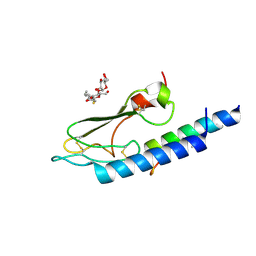 | |
1T8T
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CITRIC ACID, heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1 | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
1HV4
 
 | | CRYSTAL STRUCTURE ANALYSIS OF BAR-HEAD GOOSE HEMOGLOBIN (DEOXY FORM) | | Descriptor: | HEMOGLOBIN ALPHA-A CHAIN, HEMOGLOBIN BETA CHAIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liang, Y, Hua, Z, Liang, X, Xu, Q, Lu, G. | | Deposit date: | 2001-01-07 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bar-headed goose hemoglobin in deoxy form: the allosteric mechanism of a hemoglobin species with high oxygen affinity.
J.Mol.Biol., 313, 2001
|
|
1A9W
 
 | | HUMAN EMBRYONIC GOWER II CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), ... | | Authors: | Sutherland-Smith, A.J, Baker, H.M, Hofmann, O.M, Brittain, T, Baker, E.N. | | Deposit date: | 1998-04-11 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a human embryonic haemoglobin: the carbonmonoxy form of gower II (alpha2 epsilon2) haemoglobin at 2.9 A resolution.
J.Mol.Biol., 280, 1998
|
|
