7XZX
 
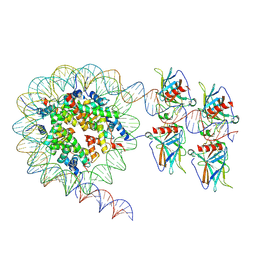 | | Cryo-EM structure of the nucleosome in complex with p53 DNA-binding domain | | Descriptor: | Cellular tumor antigen p53, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
5L9Z
 
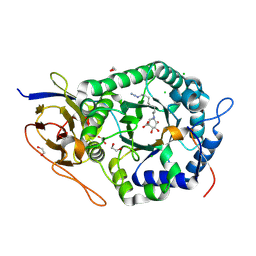 | | Crystal structure of human heparanase nucleophile mutant (E343Q), in complex with unreacted glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
6A8S
 
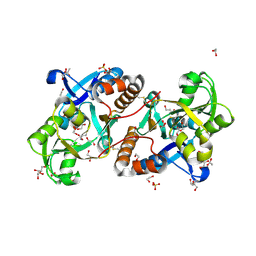 | | Crystal Structure of the putative amino acid-binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus in complex with Cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
7JP2
 
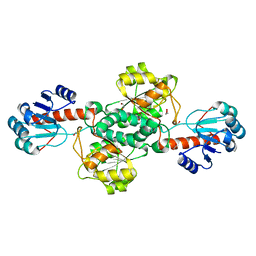 | | Crystal structure of TP0037 from Treponema pallidum, a D-lactate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-lactate dehydrogenase | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biophysical and Biochemical Characterization of TP0037, a d-Lactate Dehydrogenase, Supports an Acetogenic Energy Conservation Pathway in Treponema pallidum.
Mbio, 11, 2020
|
|
5EV9
 
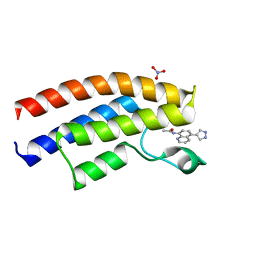 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED15 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[5-(1~{H}-pyrazol-4-yl)quinolin-8-yl]ethanamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
7TA6
 
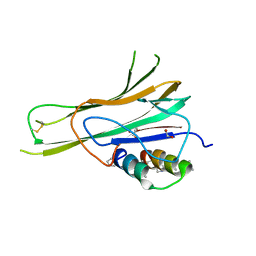 | | Trimer-to-Monomer Disruption of Tumor Necrosis Factor-alpha (TNF-alpha) by unnatural alpha/beta-peptide-1 | | Descriptor: | 1,2-ETHANEDIOL, AMINO GROUP, Alpha/Beta-peptide-1, ... | | Authors: | Niu, J, Bingman, C.A, Gellman, S.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Trimer-to-Monomer Disruption Mechanism for a Potent, Protease-Resistant Antagonist of Tumor Necrosis Factor-alpha Signaling.
J.Am.Chem.Soc., 144, 2022
|
|
3KUF
 
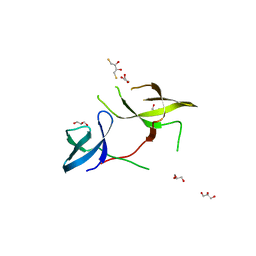 | | The Crystal Structure of the Tudor Domains from FXR1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Fragile X mental retardation syndrome-related protein 1, GLYCEROL | | Authors: | Bian, C, Guo, Y.H, Adams-Cioaba, M.A, Mackenzie, F, Kozieradzki, I, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Tudor domains from Fragile X mental retardation syndrome-related protein 1
To be Published
|
|
5GQU
 
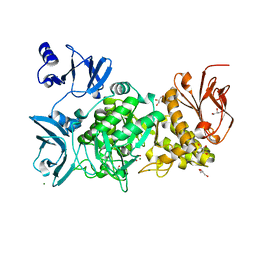 | |
6G1U
 
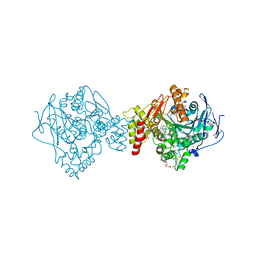 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 9-Amino-6-chloro-1,2,3,4-tetrahydro-10-methylacridin-10-ium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-10-methyl-1,2,3,4-tetrahydroacridin-10-ium-9-amine, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Increasing Polarity in Tacrine and Huprine Derivatives: Potent Anticholinesterase Agents for the Treatment of Myasthenia Gravis.
Molecules, 23, 2018
|
|
5LC8
 
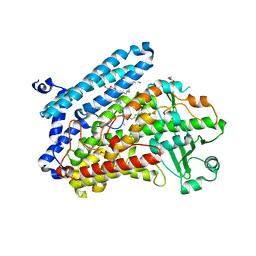 | | Crystal Structure of specific mutant from Pseudomonas aeruginosa Lipoxygenase at 1.8A resolution | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, Arachidonate 15-lipoxygenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kalms, J, Banthiya, S, Galemou Yoga, E, Kuhn, H, Scheerer, P. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Pseudomonas aeruginosa lipoxygenase Ala420Gly mutant explains the improved oxygen affinity and the altered reaction specificity.
Biochim. Biophys. Acta, 1862, 2017
|
|
7Z0P
 
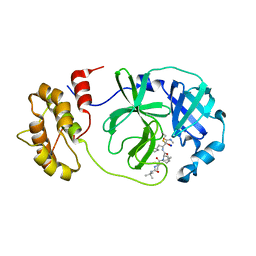 | | SARS-COV2 Main Protease in complex with inhibitor MG-131 | | Descriptor: | (1~{R},2~{S},5~{S})-3-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-~{N}-[(2~{S},3~{R})-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, SODIUM ION | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
6C5A
 
 | |
4RO3
 
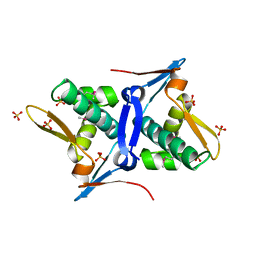 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | Hypothetical Protein, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae.
To be Published
|
|
6BUQ
 
 | |
5EWC
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED17 | | Descriptor: | NITRATE ION, Peregrin, ethyl (2~{R})-2-methyl-3-oxidanylidene-2,4-dihydro-1~{H}-quinoxaline-6-carboxylate | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
4EG9
 
 | | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
6TW6
 
 | |
5I9Z
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with danusertib (PHA739358) | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
7Z5O
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Dithionite reduced form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Mota, C, Oliveira, A.R, Klymanska, K, Pereira, I.C, Romao, M.J. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Spectroscopic and Structural Characterization of Reduced Desulfovibrio vulgaris Hildenborough W-FdhAB Reveals Stable Metal Coordination during Catalysis.
Acs Chem.Biol., 17, 2022
|
|
5CG1
 
 | | Crystal structure of E. coli FabI bound to the carbamoylated benzodiazaborine inhibitor 14b. | | Descriptor: | 1-hydroxy-2,3,1-benzodiazaborinine-2(1H)-carboxamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jordan, C.A, Vey, J.L. | | Deposit date: | 2015-07-09 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic insights into the structure-activity relationships of diazaborine enoyl-ACP reductase inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4BFA
 
 | | Crystal structure of E. coli dihydrouridine synthase C (DusC) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NITRATE ION, ... | | Authors: | Byrne, R.T, Whelan, F, Konevega, A, Aziz, N, Rodnina, M, Antson, A.A. | | Deposit date: | 2013-03-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Major Reorientation of tRNA Substrates Defines Specificity of Dihydrouridine Synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5BP3
 
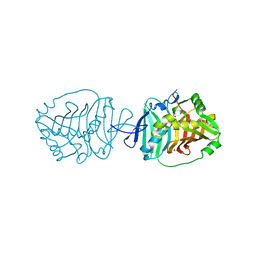 | | Dehydratase domain (DH) of a mycocerosic acid synthase-like (MAS-like) PKS, crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Mycocerosic acid synthase-like polyketide synthase | | Authors: | Herbst, D.A, Jakob, P.R, Zaehringer, F, Maier, T. | | Deposit date: | 2015-05-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mycocerosic acid synthase exemplifies the architecture of reducing polyketide synthases.
Nature, 531, 2016
|
|
2W9H
 
 | |
6MBK
 
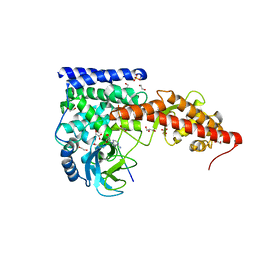 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, First P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6CAZ
 
 | |
