4LS3
 
 | | THE crystal STRUCTURE OF HELICOBACTER PYLORI CEUE(HP1561)/NI-HIS COMPL | | Descriptor: | HISTIDINE, NICKEL (II) ION, Nickel (III) ABC transporter, ... | | Authors: | Salamina, M, Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
2OXY
 
 | | Protein kinase CK2 in complex with tetrabromobenzoimidazole derivatives K17, K22 and K32 | | Descriptor: | 4,5,6,7-TETRABROMO-BENZIMIDAZOLE, Casein kinase II subunit alpha | | Authors: | Battistutta, R, Zanotti, G, Cendron, L. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | The ATP-Binding Site of Protein Kinase CK2 Holds a Positive Electrostatic Area and Conserved Water Molecules.
Chembiochem, 8, 2007
|
|
2OXD
 
 | | Protein kinase CK2 in complex with tetrabromobenzoimidazole K17, K22 and K32 inhibitors | | Descriptor: | 4,5,6,7-TETRABROMO-1H,3H-BENZIMIDAZOL-2-ONE, Casein kinase II subunit alpha | | Authors: | Battistutta, R, Zanotti, G, Cendron, L. | | Deposit date: | 2007-02-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ATP-Binding Site of Protein Kinase CK2 Holds a Positive Electrostatic Area and Conserved Water Molecules.
Chembiochem, 8, 2007
|
|
3RQ1
 
 | | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula | | Descriptor: | 2-OXOGLUTARIC ACID, Aminotransferase class I and II, CHLORIDE ION, ... | | Authors: | Kim, Y, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula
To be Published
|
|
2OXX
 
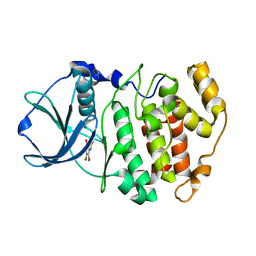 | | Protein kinase CK2 in complex with tetrabromobenzoimidazole derivatives K17, K22 and K32 | | Descriptor: | 4,5,6,7-TETRABROMO-1H,3H-BENZIMIDAZOL-2-THIONE, Casein kinase II subunit alpha | | Authors: | Battistutta, R, Zanotti, G, Cendron, L. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ATP-Binding Site of Protein Kinase CK2 Holds a Positive Electrostatic Area and Conserved Water Molecules.
Chembiochem, 8, 2007
|
|
1OM1
 
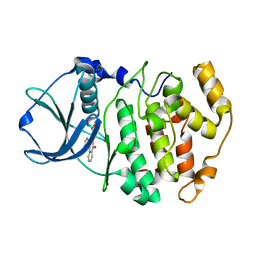 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
4INO
 
 | |
4INP
 
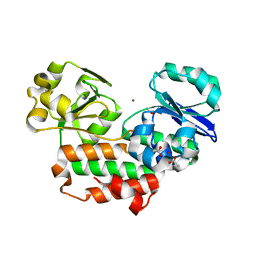 | | The crystal structure of Helicobacter pylori Ceue (HP1561) with Ni(II) bound | | Descriptor: | ACETATE ION, Iron (III) ABC transporter, periplasmic iron-binding protein, ... | | Authors: | Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-01-05 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
1TTJ
 
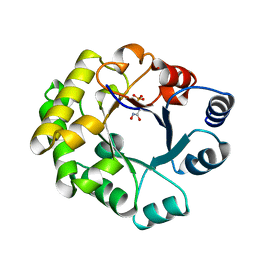 | |
1TTI
 
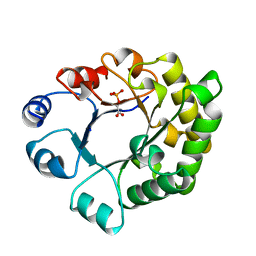 | |
3ISL
 
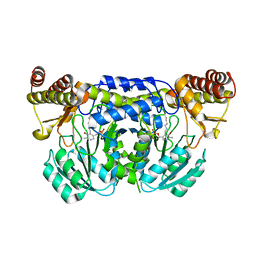 | | Crystal structure of ureidoglycine-glyoxylate aminotransferase (pucG) from Bacillus subtilis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Purine catabolism protein pucG | | Authors: | Costa, R, Cendron, L, Ramazzina, I, Berni, R, Peracchi, A, Percudani, R, Zanotti, G. | | Deposit date: | 2009-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Amino acids from purines in GUT bacteria
To be Published
|
|
1TRI
 
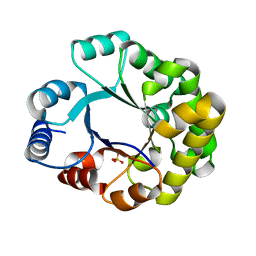 | |
3BHV
 
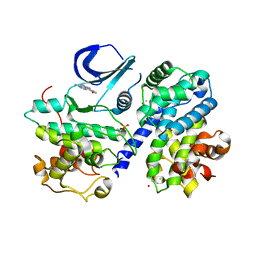 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor variolin B | | Descriptor: | 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
7E9B
 
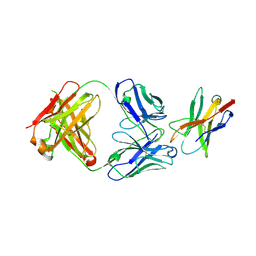 | |
3R6B
 
 | | Crystal Structure of Thrombospondin-1 TSR Domains 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1 | | Authors: | Page, R.C, Klenotic, P.A, Misra, S, Silverstein, R.L. | | Deposit date: | 2011-03-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, purification and structural characterization of functionally replete thrombospondin-1 type 1 repeats in a bacterial expression system.
Protein Expr.Purif., 80, 2011
|
|
3BHT
 
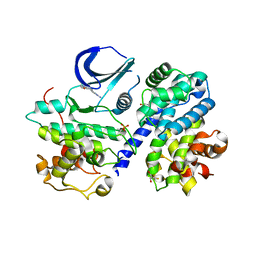 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor meriolin 3 | | Descriptor: | 4-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
2XH1
 
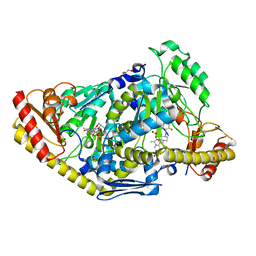 | | Crystal structure of human KAT II-inhibitor complex | | Descriptor: | (3S)-10-(4-AMINOPIPERAZIN-1-YL)-9-FLUORO-7-HYDROXY-3-METHYL-2,3-DIHYDRO-8H-[1,4]OXAZINO[2,3,4-IJ]QUINOLINE-6-CARBOXYLATE, GLYCEROL, IODIDE ION, ... | | Authors: | Rossi, F, Casazza, V, Garavaglia, S, Sathyasaikumar, K.V, Schwarcz, R, Kojima, S.I, Okuwaki, K, Ono, S.I, Kajii, Y, Rizzi, M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure-Based Selective Targeting of the Pyridoxal 5'-Phsosphate Dependent Enzyme Kynurenine Aminotransferase II for Cognitive Enhancement
J.Med.Chem., 53, 2010
|
|
7D5L
 
 | | Discovery of BMS-986144, a Third Generation, Pan Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection | | Descriptor: | NS3/4A Protease, ZINC ION, [1,1,1-tris(fluoranyl)-2-methyl-propan-2-yl] ~{N}-[(1~{S},4~{R},6~{S},7~{Z},11~{R},13~{R},14~{S},18~{R})-13-ethyl-18-(7-fluoranyl-6-methoxy-isoquinolin-1-yl)oxy-11-methyl-4-[(1-methylcyclopropyl)sulfonylcarbamoyl]-2,15-bis(oxidanylidene)-3,16-diazatricyclo[14.3.0.0^{4,6}]nonadec-7-en-14-yl]carbamate | | Authors: | Ghosh, K, Anumula, R, Kumar, A. | | Deposit date: | 2020-09-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 63, 2020
|
|
4APN
 
 | | Structure of TR from Leishmania infantum in complex with a diarylpirrole-based inhibitor | | Descriptor: | 4-[[1-(4-ethylphenyl)-2-methyl-5-(4-methylsulfanylphenyl)pyrrol-3-yl]methyl]thiomorpholine, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G, Biava, M. | | Deposit date: | 2012-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Leishmania Infantum Trypanothione Reductase by Azole-Based Compounds: A Comparative Analysis with its Physiological Substrate by X-Ray Crystallography.
Chemmedchem, 8, 2013
|
|
2WCI
 
 | | Structure of E. coli monothiol glutaredoxin GRX4 homodimer | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTAREDOXIN-4, GLUTATHIONE, ... | | Authors: | Iwema, T, Picchiocci, A, Traore, D.A.K, Ferrer, J.-L, Chauvat, F, Jacquamet, L. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Delivery of the Intact [Fe2S2] Cluster by Monothiol Glutaredoxin.
Biochemistry, 48, 2009
|
|
3LY1
 
 | |
4ATO
 
 | | New insights into the mechanism of bacterial Type III toxin-antitoxin systems: selective toxin inhibition by a non-coding RNA pseudoknot | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, TOXI, TOXN | | Authors: | Short, F.L, Pei, X.Y, Blower, T.R, Ong, S.L, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2012-05-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity and Self-Assembly in the Control of a Bacterial Toxin by an Antitoxic Noncoding RNA Pseudoknot.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VFE
 
 | | Virtual Screening and X-Ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group | | Descriptor: | 4-{[(3R)-3-{[(7-methoxynaphthalen-2-yl)sulfonyl](thiophen-3-ylmethyl)amino}-2-oxopyrrolidin-1-yl]methyl}thiophene-2-carboximidamide, Kallikrein-6 | | Authors: | Chen, X, Zhang, Y, Xia, T, Wang, R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Acs Med.Chem.Lett., 3, 2012
|
|
3JZK
 
 | | crystal structure of MDM2 with chromenotriazolopyrimidine 1 | | Descriptor: | (6R,7S)-6,7-bis(4-bromophenyl)-7,11-dihydro-6H-chromeno[4,3-d][1,2,4]triazolo[1,5-a]pyrimidine, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction.
J.Med.Chem., 52, 2009
|
|
3V1R
 
 | | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of XMRV with inhibitor beta-thujaplicinol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, ... | | Authors: | Zhou, D, Wlodawer, A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of xenotropic murine leukemia-virus related virus.
J.Struct.Biol., 177, 2012
|
|
