6DVV
 
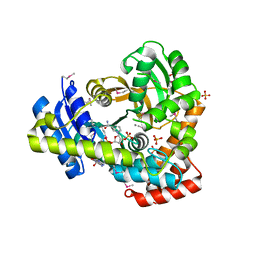 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
2V5I
 
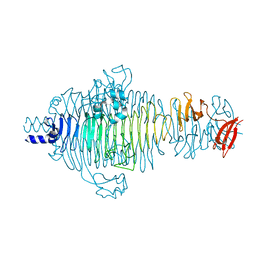 | | Structure of the receptor-binding protein of bacteriophage Det7: a podoviral tailspike in a myovirus | | Descriptor: | SALMONELLA TYPHIMURIUM DB7155 BACTERIOPHAGE DET7 TAILSPIKE, SODIUM ION | | Authors: | Walter, M, Fiedler, C, Grassl, R, Biebl, M, Rachel, R, Hermo-Parrado, X.L, Llamas-Saiz, A.L, Seckler, R, Miller, S, van Raaij, M.J. | | Deposit date: | 2007-07-05 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Receptor-Binding Protein of Bacteriophage Det7: A Podoviral Tail Spike in a Myovirus.
J.Virol., 82, 2008
|
|
3HM7
 
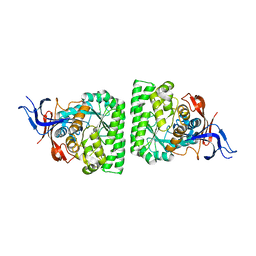 | | Crystal structure of allantoinase from Bacillus halodurans C-125 | | Descriptor: | Allantoinase, ZINC ION | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Miller, S, Wasserman, S.R, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Allantoinase from Bacillus Halodurans
To be Published
|
|
2LDO
 
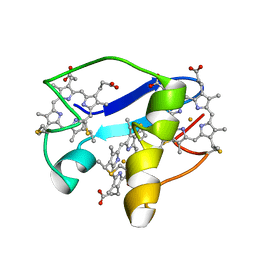 | | Solution structure of triheme cytochrome PpcA from Geobacter sulfurreducens reveals the structural origin of the redox-Bohr effect | | Descriptor: | Cytochrome c3, HEME C | | Authors: | Morgado, L, Paixao, V.B, Bruix, M, Salgueiro, C.A. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | Revealing the structural origin of the redox-Bohr effect: the first solution structure of a cytochrome from Geobacter sulfurreducens.
Biochem.J., 441, 2012
|
|
2PLM
 
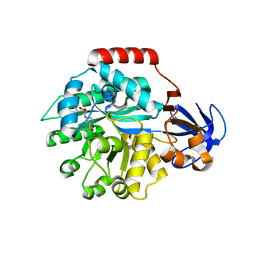 | | Crystal structure of the protein TM0936 from Thermotoga maritima complexed with ZN and S-inosylhomocysteine | | Descriptor: | (2S)-2-AMINO-4-({[(2S,3S,4R,5R)-3,4-DIHYDROXY-5-(6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)TETRAHYDROFURAN-2-YL]METHYL}THIO)BUTANOIC ACID, Uncharacterized protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Hermann, J.C, Marti-Arbona, R, Shoichet, B.K, Raushel, F.M, Almo, S.C. | | Deposit date: | 2007-04-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based activity prediction for an enzyme of unknown function
Nature, 448, 2007
|
|
7BKB
 
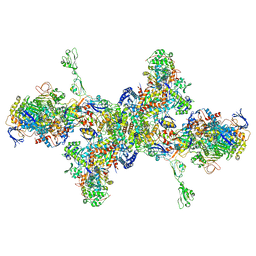 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (hexameric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
2JJP
 
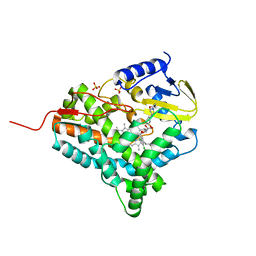 | | Structure of cytochrome P450 EryK in complex with inhibitor ketoconazole (KC) | | Descriptor: | 1-ACETYL-4-(4-{[(2S,4R)-2-(2,4-DICHLOROPHENYL)-2-(1H-IMIDAZOL-1-YLMETHYL)-1,3-DIOXOLAN-4-YL]METHOXY}PHENYL)PIPERAZINE, CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Azole Drugs Trap Cytochrome P450 Eryk in Alternative Conformational States.
Biochemistry, 49, 2010
|
|
2PG6
 
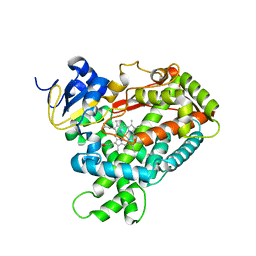 | | Crystal Structure of Human Microsomal P450 2A6 L240C/N297Q | | Descriptor: | Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sansen, S, Hsu, M.H, Stout, C.D, Johnson, E.F. | | Deposit date: | 2007-04-06 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants.
Arch.Biochem.Biophys., 464, 2007
|
|
2PMQ
 
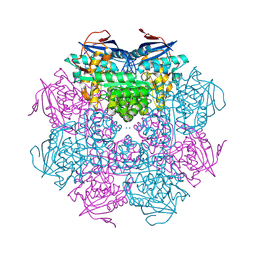 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
3LT5
 
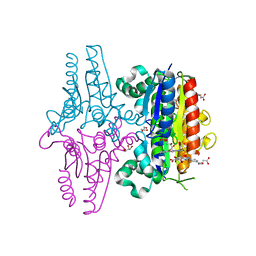 | | X-ray Crystallographic structure of a Pseudomonas Aeruginosa Azoreductase in complex with balsalazide | | Descriptor: | (3E)-3-({4-[(2-carboxyethyl)carbamoyl]phenyl}hydrazono)-6-oxocyclohexa-1,4-diene-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Laurieri, N, Westwood, I, Wang, C.-J, Lowe, E, Sim, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Azoreduction
J.Mol.Biol., 400, 2010
|
|
6GDF
 
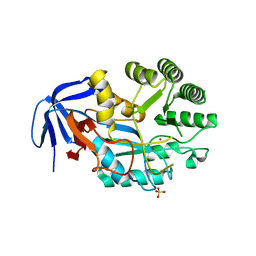 | | DIHYDROOROTASE FROM AQUIFEX AEOLICUS UNDER 600 BAR OF HYDROSTATIC PRESSURE | | Descriptor: | Dihydroorotase, SULFATE ION, ZINC ION | | Authors: | Prange, T, Girard, E, Herve, G, Evans, D.R. | | Deposit date: | 2018-04-23 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pressure-induced activation of latent dihydroorotase from Aquifex aeolicus as revealed by high pressure protein crystallography.
Febs J., 286, 2019
|
|
6GEO
 
 | |
6GEQ
 
 | |
5Z21
 
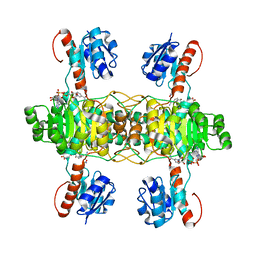 | | The ternary structure of D-lactate dehydrogenase from Fusobacterium nucleatum with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase, OXAMIC ACID | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
7E3U
 
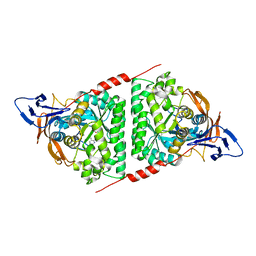 | | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-AU | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Yang, Y.C, Luo, R.H, Huang, Y.H, Huang, C.Y, Lin, E.S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil.
Bioinorg Chem Appl, 2022, 2022
|
|
3KTU
 
 | |
2XMN
 
 | | High resolution snapshots of defined TolC open states present an iris- like movement of periplasmic entrance helices | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Outer membrane protein TolC | | Authors: | Pei, X.Y, Koronakis, E, Hughes, C, Koronakis, V. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of sequential open states in a symmetrical opening transition of the TolC exit duct.
Proc. Natl. Acad. Sci. U.S.A., 108, 2011
|
|
2OHO
 
 | |
2JJO
 
 | | Structure of cytochrome P450 EryK in complex with its natural substrate erD | | Descriptor: | CYTOCHROME P450 113A1, Erythromycin D, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2JFU
 
 | |
2PG7
 
 | | Crystal Structure of Human Microsomal P450 2A6 N297Q/I300V | | Descriptor: | Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sansen, S, Hsu, M.H, Stout, C.D, Johnson, E.F. | | Deposit date: | 2007-04-06 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants.
Arch.Biochem.Biophys., 464, 2007
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | Descriptor: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | Authors: | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
2YKF
 
 | | Sensor region of a sensor histidine kinase | | Descriptor: | BROMIDE ION, PROBABLE SENSOR HISTIDINE KINASE PDTAS, SULFATE ION | | Authors: | Preu, J, Panjikar, S, Morth, P, Jaiswal, R, Karunakar, P, Tucker, P.A. | | Deposit date: | 2011-05-26 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Sensor Region of the Ubiquitous Cytosolic Sensor Kinase, Pdtas, Contains Pas and Gaf Domain Sensing Modules.
J.Struct.Biol., 177, 2012
|
|
2JFX
 
 | |
