2PNH
 
 | | Escherichia coli PriB E39A variant | | Descriptor: | Primosomal replication protein n | | Authors: | Lopper, M.E, Keck, J.L. | | Deposit date: | 2007-04-24 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A hand-off mechanism for primosome assembly in replication restart.
Mol.Cell, 26, 2007
|
|
4YV6
 
 | |
1JUM
 
 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to the natural drug berberine | | Descriptor: | BERBERINE, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
5D1W
 
 | | Crystal structure of Mycobacterium tuberculosis Rv3249c transcriptional regulator. | | Descriptor: | PALMITIC ACID, Rv3249c transcriptional regulator | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
2KHL
 
 | |
2OQ4
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(*C*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*G*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Grollman, A.P, Shoahm, G. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrates and mutations of critical residues
To be Published
|
|
2OPF
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (R252A) in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*G)-3', 5'-D(P*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharov, D.O, Grollman, A.P, Shoham, G. | | Deposit date: | 2007-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrates and mutations of critical residues
To be Published
|
|
5D1R
 
 | | Crystal structure of Mycobacterium tuberculosis Rv1816 transcriptional regulator. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Rv1816 transcriptional regulator, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
3GN5
 
 | | Structure of the E. coli protein MqsA (YgiT/b3021) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator MQSA (YGIT/b3021), ZINC ION | | Authors: | Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | Deposit date: | 2009-03-16 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
6KVN
 
 | |
5E5A
 
 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | Descriptor: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | Authors: | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
1IG6
 
 | | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES | | Descriptor: | MODULATOR RECOGNITION FACTOR 2 | | Authors: | Lin, D, Tsui, V, Case, D, Yuan, Y.C, Chen, Y. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES
To be Published
|
|
6OD7
 
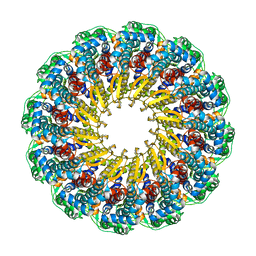 | | Herpes simplex virus type 1 (HSV-1) pUL6 portal protein, dodecameric complex | | Descriptor: | Portal protein | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
5HXB
 
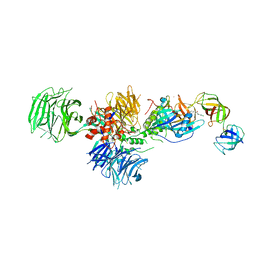 | | Cereblon in complex with DDB1, CC-885, and GSPT1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Chamberlain, P.P, Matyskiela, M, Pagarigan, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Nature, 535, 2016
|
|
6LCM
 
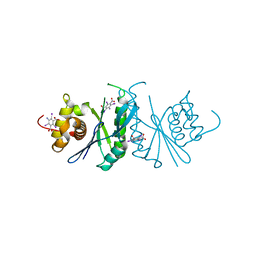 | | Crystal structure of chloroplast resolvase ZmMOC1 with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, ZmMoc1 | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6L0C
 
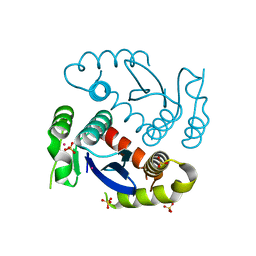 | |
6DKA
 
 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | CYANAMIDE, DNA damage-inducible protein, SULFATE ION, ... | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
1PCZ
 
 | | STRUCTURE OF TATA-BINDING PROTEIN | | Descriptor: | TATA-BINDING PROTEIN | | Authors: | Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1996-10-04 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a hyperthermophilic archaeal TATA-box binding protein.
J.Mol.Biol., 264, 1996
|
|
5EXT
 
 | |
5EXP
 
 | | AAA+ domain of FleQ from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Transcriptional regulator FleQ | | Authors: | Navarro, M.V.A.S, Sondermann, H, Matsuyama, B.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ODG
 
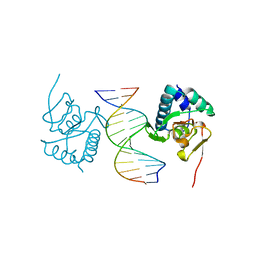 | | Crystal structure of Smad3-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 3, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5EXS
 
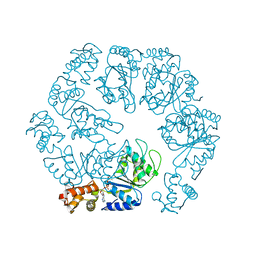 | |
4B0A
 
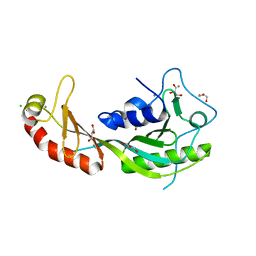 | | The high-resolution structure of yTBP-yTAF1 identifies conserved and competing interaction surfaces in transcriptional activation | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Anandapadamanaban, M, Andresen, C, Siponen, M, Kokubo, T, Ikura, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High-Resolution Structure of TBP with Taf1 Reveals Anchoring Patterns in Transcriptional Regulation
Nat.Struct.Mol.Biol., 20, 2013
|
|
7XL9
 
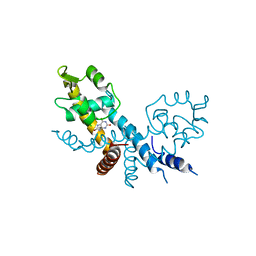 | | The structure of HucR with urate | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family, ... | | Authors: | Park, S.Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The structure of Deinococcus radiodurans transcriptional regulator HucR retold with the urate bound.
Biochem.Biophys.Res.Commun., 615, 2022
|
|
6FHS
 
 | | CryoEM Structure of INO80core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Arp5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-15 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.754 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
