4MT2
 
 | |
2IMM
 
 | |
1RAS
 
 | | CRYSTAL STRUCTURE OF A FLUORESCENT DERIVATIVE OF RNASE A | | Descriptor: | 5-(1-SULFONAPHTHYL)-ACETYLAMINO-ETHYLAMINE, RIBONUCLEASE A | | Authors: | Baudet-Nessler, S, Jullien, M, Crosio, M.-P, Janin, J. | | Deposit date: | 1993-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a fluorescent derivative of RNase A.
Biochemistry, 32, 1993
|
|
1RAR
 
 | | CRYSTAL STRUCTURE OF A FLUORESCENT DERIVATIVE OF RNASE A | | Descriptor: | 5-(1-SULFONAPHTHYL)-ACETYLAMINO-ETHYLAMINE, CHLORIDE ION, RIBONUCLEASE A | | Authors: | Baudet-Nessler, S, Jullien, M, Crosio, M.-P, Janin, J. | | Deposit date: | 1993-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a fluorescent derivative of RNase A.
Biochemistry, 32, 1993
|
|
1BPN
 
 | |
1BLL
 
 | |
1BPM
 
 | |
1CRL
 
 | |
1ALK
 
 | |
1AZR
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA ZINC AZURIN MUTANT ASP47ASP AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION | | Authors: | Sjolin, L, Tsai, Lc, Langer, V, Pascher, T, Karlsson, G, Nordling, M, Nar, H. | | Deposit date: | 1993-03-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Pseudomonas aeruginosai zinc azurin mutant Asn47Asp at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1FPV
 
 | |
1AVE
 
 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE APO-AVIDIN IN RELATION TO ITS THERMAL STABILITY PROPERTIES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AVIDIN | | Authors: | Pugliese, L, Coda, A, Malcovati, M, Bolognesi, M. | | Deposit date: | 1993-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of apo-avidin from hen egg-white.
J.Mol.Biol., 235, 1994
|
|
1HIQ
 
 | |
1RDH
 
 | |
1AVD
 
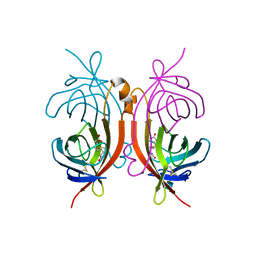 | | THREE-DIMENSIONAL STRUCTURE OF THE TETRAGONAL CRYSTAL FORM OF EGG-WHITE AVIDIN IN ITS FUNCTIONAL COMPLEX WITH BIOTIN AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AVIDIN, BIOTIN | | Authors: | Pugliese, L, Coda, A, Malcovati, M, Bolognesi, M. | | Deposit date: | 1993-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of the tetragonal crystal form of egg-white avidin in its functional complex with biotin at 2.7 A resolution.
J.Mol.Biol., 231, 1993
|
|
1SPS
 
 | |
1SPR
 
 | |
2ABD
 
 | |
1NNB
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1NNA
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1WAT
 
 | |
1WAS
 
 | |
1DCB
 
 | |
1GLV
 
 | |
1NBV
 
 | |
