5TXN
 
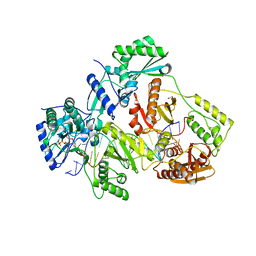 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
8AEO
 
 | |
8AER
 
 | |
405D
 
 | |
7X9S
 
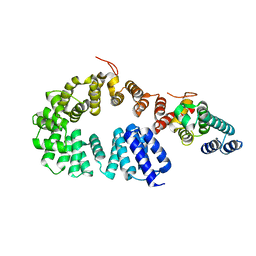 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
6UJF
 
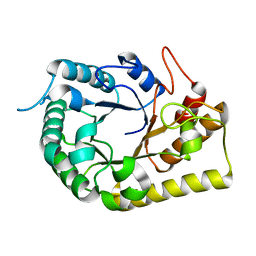 | | Crystal structure of the Clostridial cellulose synthase subunit Z (CcsZ) from Clostridioides difficile | | Descriptor: | Endoglucanase, beta-D-glucopyranose | | Authors: | Scott, W, Lowrance, B, Anderson, A.C, Weadge, J.T. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of the Clostridial cellulose synthase and characterization of the cognate glycosyl hydrolase, CcsZ.
Plos One, 15, 2020
|
|
1NU4
 
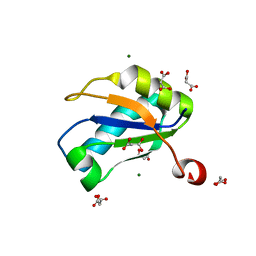 | | U1A RNA binding domain at 1.8 angstrom resolution reveals a pre-organized C-terminal helix | | Descriptor: | MAGNESIUM ION, MALONIC ACID, U1A RNA binding domain | | Authors: | Rupert, P.B, Xiao, H, Ferre-D'Amare, A.R. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | U1A RNA-binding domain at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6G4B
 
 | |
5EYK
 
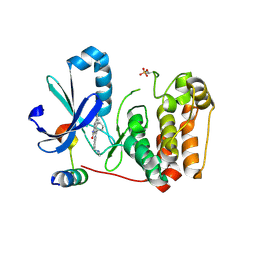 | | CRYSTAL STRUCTURE OF AURORA B IN COMPLEX WITH BI 847325 | | Descriptor: | 3-[(3~{Z})-3-[[[4-[(dimethylamino)methyl]phenyl]amino]-phenyl-methylidene]-2-oxidanylidene-1~{H}-indol-6-yl]-~{N}-ethyl-prop-2-ynamide, Aurora kinase B-A, Inner centromere protein A | | Authors: | Bader, G, Zoephel, A. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-17 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
7RDT
 
 | | Structure of human NTHL1 - linker 1 chimera | | Descriptor: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | Authors: | Carroll, B.L, Doublie, S. | | Deposit date: | 2021-07-11 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
8A92
 
 | | p53-Y220C Core Domain in Complex with a Bromo-trifluoro-pyrazole-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromanyl-5-(trifluoromethyl)-1H-pyrazol-3-amine, Cellular tumor antigen p53, ... | | Authors: | Stahlecker, J, Braun, M.B, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C.
Rsc Med Chem, 13, 2022
|
|
8UH1
 
 | |
6UN9
 
 | | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Seetharaman, J, Kolev, M, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2019-10-11 | | Release date: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25
To Be Published
|
|
6UD1
 
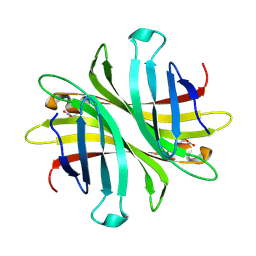 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | CHLORIDE ION, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
5CIB
 
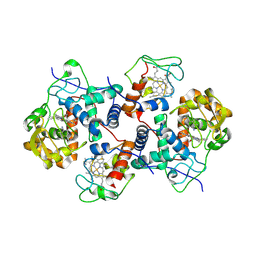 | | Complex of yeast cytochrome c peroxidase (W191G) bound to 2,4-dimethylaniline with iso-1 cytochrome c | | Descriptor: | 2,4-dimethylaniline, Cytochrome c iso-1, Cytochrome c peroxidase, ... | | Authors: | Crane, B.R, Payne, T.M. | | Deposit date: | 2015-07-11 | | Release date: | 2016-08-03 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Constraints on the Radical Cation Center of Cytochrome c Peroxidase for Electron Transfer from Cytochrome c.
Biochemistry, 55, 2016
|
|
7LVQ
 
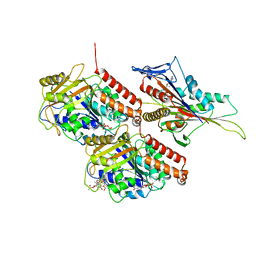 | | KIF14[391-743] - AMP-PNP closed state class in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF14, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Paydar, M, Dhakal, S, Kwok, B, Sosa, H. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14.
Nat Commun, 12, 2021
|
|
5MJC
 
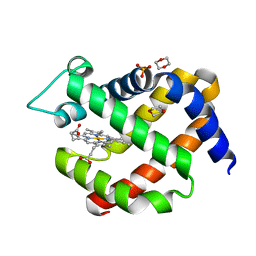 | | metNeuroglobin under oxygen at 50 bar | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, OXYGEN MOLECULE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
5FO1
 
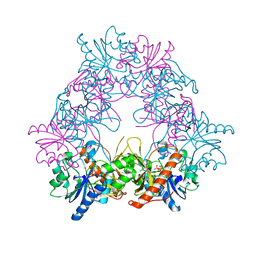 | |
5U8V
 
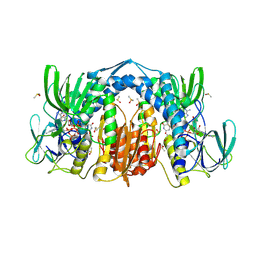 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa bound to NAD+ | | Descriptor: | DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
7K9M
 
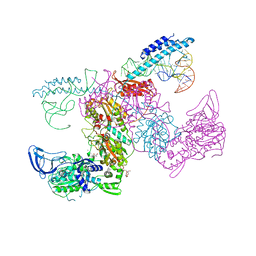 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
6BWY
 
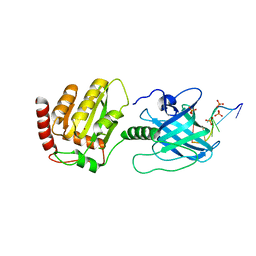 | | DNA substrate selection by APOBEC3G | | Descriptor: | DNA (30-MER), PHOSPHATE ION, Protection of telomeres protein 1, ... | | Authors: | Ziegler, S.J, Buzovetsky, O. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into DNA substrate selection by APOBEC3G from structural, biochemical, and functional studies.
PLoS ONE, 13, 2018
|
|
1KNO
 
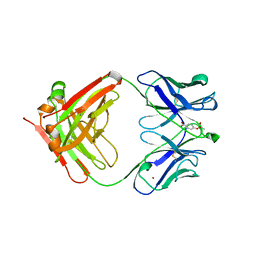 | |
436D
 
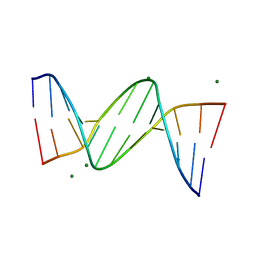 | |
4RU5
 
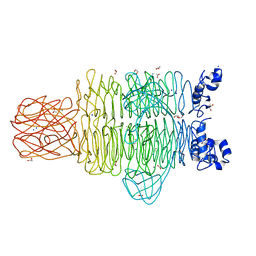 | | Crystal Structure of the Pseudomonas phage phi297 tailspike gp61 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Browning, C, Sycheva, L.V, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
5C7T
 
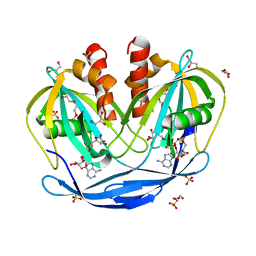 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase in complex with ADP-ribose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5-DIPHOSPHORIBOSE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
