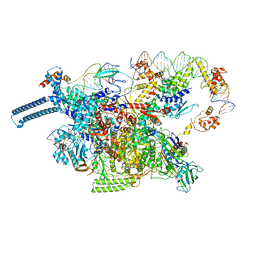
8DY9
 
 | |
3PED
 
 | |
3L03
 
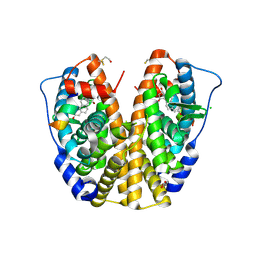 | | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II peptide and Estetrol (Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol) | | Descriptor: | (14beta,15alpha,16alpha,17alpha)-estra-1,3,5(10)-triene-3,15,16,17-tetrol, CHLORIDE ION, Estrogen receptor, ... | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain
in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II
peptide and Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol
To be Published
|
|
3PEC
 
 | |
5NJB
 
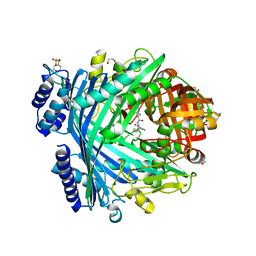 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5OBH
 
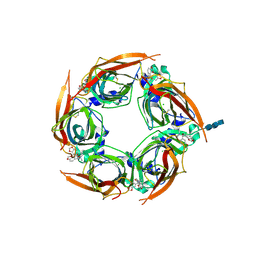 | | Crystal structure of glycine binding protein in complex with bicuculline | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dawson, A, Hunter, W.N, Jones, M. | | Deposit date: | 2017-06-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Structural Rationale for N-Methylbicuculline Acting as a Promiscuous Competitive Antagonist of Inhibitory Pentameric Ligand-Gated Ion Channels.
Chembiochem, 21, 2020
|
|
8JE4
 
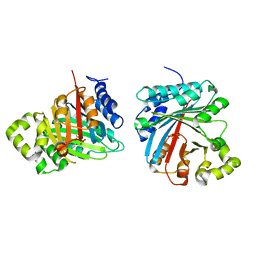 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | Authors: | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
3OV8
 
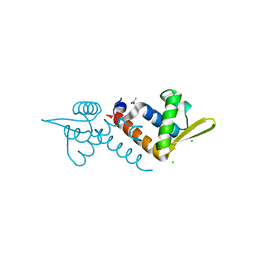 | | Crystal structure of AF1382 from Archaeoglobus fulgidus, High resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, Protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-09-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5DTR
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD5 [N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
3KCP
 
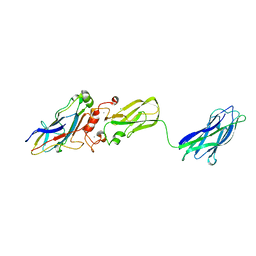 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
3O83
 
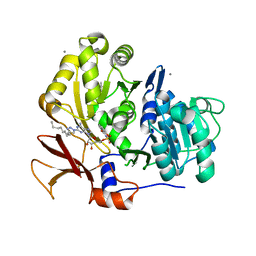 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O3K
 
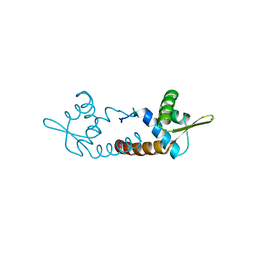 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Xu, H, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1KPE
 
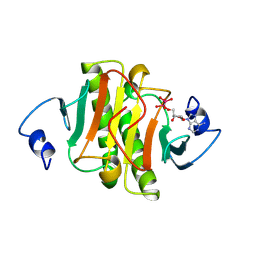 | | PKCI-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE-5'-DITUNGSTATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
3O84
 
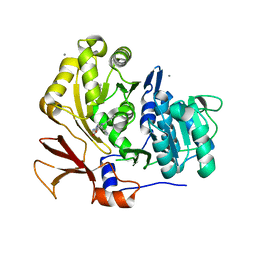 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O82
 
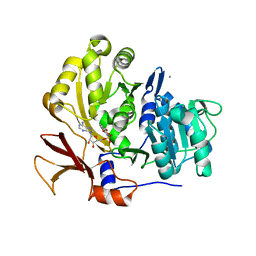 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine | | Descriptor: | 5'-O-{[(2,3-dihydroxyphenyl)carbonyl]sulfamoyl}adenosine, CALCIUM ION, Peptide arylation enzyme | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
1B77
 
 | |
1PD7
 
 | | Extended SID of Mad1 bound to the PAH2 domain of mSin3B | | Descriptor: | Mad1, Sin3b protein | | Authors: | Van Ingen, H, Lasonder, E, Jansen, J.F, Kaan, A.M, Spronk, C.A, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2003-05-19 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Extension of the binding motif of the sin3 interacting domain of the mad family proteins(,).
Biochemistry, 43, 2004
|
|
1AV5
 
 | | PKCI-SUBSTRATE ANALOG | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
3C87
 
 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of enterobactin | | Descriptor: | Enterochelin esterase, PHOSPHATE ION, SODIUM ION | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Raymond, K, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-11 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
3C8H
 
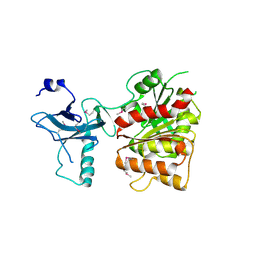 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of 2,3-Di-hydroxy-N-benzoyl-serine | | Descriptor: | Enterochelin esterase | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Holzle, D, Raymond, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
1RY1
 
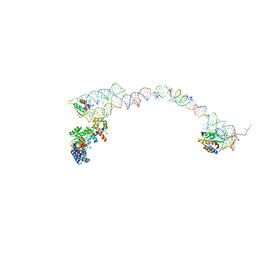 | | Structure of the signal recognition particle interacting with the elongation-arrested ribosome | | Descriptor: | SRP Alu domain, SRP RNA, SRP S domain, ... | | Authors: | Halic, M, Becker, T, Pool, M.R, Spahn, C.M, Grassucci, R.A, Frank, J, Beckmann, R. | | Deposit date: | 2003-12-19 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the signal recognition particle interacting with the elongation-arrested ribosome
Nature, 427, 2004
|
|
8A1N
 
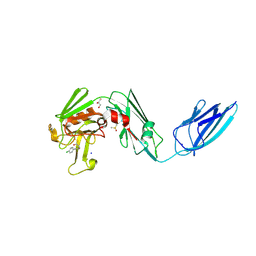 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with fumaryl amide analogue 13 | | Descriptor: | (Z)-N-(4-chlorophenyl)-4-oxidanylidene-but-2-enamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
8A1M
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with maleimide analogue 4 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-fluoranyl-5-methylsulfonyl-phenyl)pyrrolidine-2,5-dione, CHLORIDE ION, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
8A1K
 
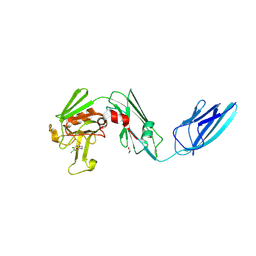 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with ebsulfur analogue 15 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-bis(chloranyl)-N-(2-hydroxyethyl)-2-sulfanyl-benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
8A1O
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with acrylamide analogue 8 | | Descriptor: | (E)-3-chloranyl-3-[(2-chlorophenyl)methylsulfonyl]-N-(5-methoxypyridin-2-yl)prop-2-enamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
