1EE3
 
 | | Cadmium-substituted bovine pancreatic carboxypeptidase A (alfa-form) at pH 7.5 and 2 mM chloride in monoclinic crystal form | | Descriptor: | CADMIUM ION, PROTEIN (CARBOXYPEPTIDASE A) | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-01-30 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ELM
 
 | | CADMIUM-SUBSTITUTED BOVINE PACREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 5.5 AND 2 MM CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ELL
 
 | | CADMIUM-SUBSTITUTED BOVINE PANCREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 7.5 AND 0.25 M CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
7EV1
 
 | | Crystal structure of LI-Cadherin EC1-2 | | Descriptor: | ACETATE ION, CALCIUM ION, Cadherin-17, ... | | Authors: | Caaveiro, J.M.M, Yui, A, Tsumoto, K. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mechanism of dimerization and structural features of human LI-cadherin.
J.Biol.Chem., 297, 2021
|
|
3Q2W
 
 | | Crystal structure of mouse N-cadherin ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
2O72
 
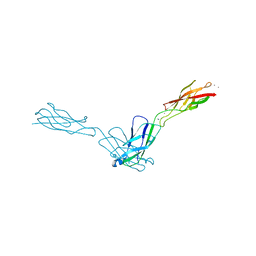 | | Crystal Structure Analysis of human E-cadherin (1-213) | | Descriptor: | CALCIUM ION, Epithelial-cadherin | | Authors: | Parisini, E, Wang, J.-H. | | Deposit date: | 2006-12-09 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human E-cadherin Domains 1 and 2, and Comparison with other Cadherins in the Context of Adhesion Mechanism
J.Mol.Biol., 373, 2007
|
|
2QVF
 
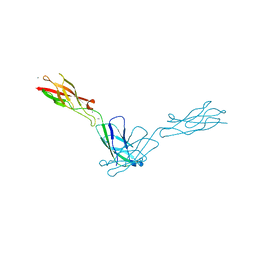 | | mouse E-cadherin domains 1,2 | | Descriptor: | CALCIUM ION, Epithelial-cadherin (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) | | Authors: | Carroll, K.J. | | Deposit date: | 2007-08-08 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E-cadherin domains 1,2
To be Published
|
|
3K5R
 
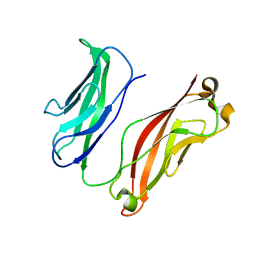 | |
3K6D
 
 | |
5JYM
 
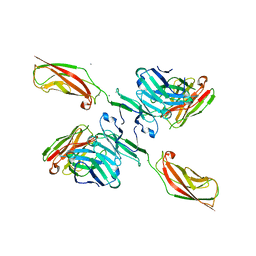 | | Human P-cadherin EC12 with scFv TSP11 bound | | Descriptor: | Antibody scFv TSP11, CALCIUM ION, Cadherin-3 | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin
Sci Rep, 7, 2017
|
|
6ZTR
 
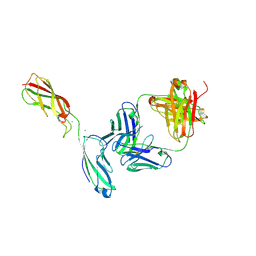 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 in complex with human P-Cadherin(108-324) | | Descriptor: | CALCIUM ION, CQY684 Fab heavy-chain, CQY684 Fab light-chain, ... | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTB
 
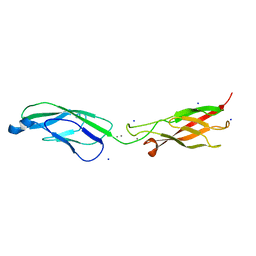 | | Crystal Structure of human P-Cadherin EC1_EC2 | | Descriptor: | CALCIUM ION, Cadherin-3, SODIUM ION | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
5JYL
 
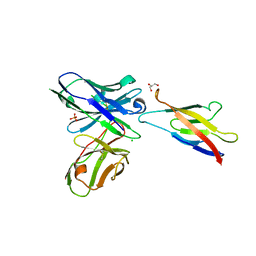 | | Human P-cadherin MEC1 with scFv TSP7 bound | | Descriptor: | CHLORIDE ION, Cadherin-3, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin
Sci Rep, 7, 2017
|
|
3QRB
 
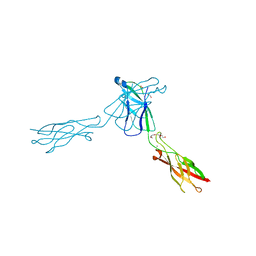 | | crystal structure of E-cadherin EC1-2 P5A P6A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-1, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2011-02-17 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular design principles underlying beta-strand swapping in the adhesive dimerization of cadherins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4AQE
 
 | | CRYSTAL STRUCTURE OF DEAFNESS ASSOCIATED MUTANT MOUSE CADHERIN-23 EC1- 2S70P AND PROTOCADHERIN-15 EC1-2 FORM I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CADHERIN-23, CALCIUM ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2012-04-16 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Force-Conveying Cadherin Bond Essential for Inner-Ear Mechanotransduction
Nature, 492, 2012
|
|
4AQA
 
 | | CRYSTAL STRUCTURE OF DEAFNESS ASSOCIATED MUTANT MOUSE CADHERIN-23 EC1- 2D124G AND PROTOCADHERIN-15 EC1-2 FORM I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CADHERIN-23, CALCIUM ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2012-04-15 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a Force-Conveying Cadherin Bond Essential for Inner-Ear Mechanotransduction
Nature, 492, 2012
|
|
4APX
 
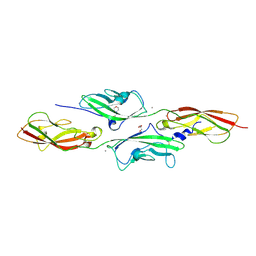 | | CRYSTAL STRUCTURE OF MOUSE CADHERIN-23 EC1-2 AND PROTOCADHERIN-15 EC1- 2 FORM I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CADHERIN-23, CALCIUM ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2012-04-07 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a Force-Conveying Cadherin Bond Essential for Inner-Ear Mechanotransduction
Nature, 492, 2012
|
|
2A62
 
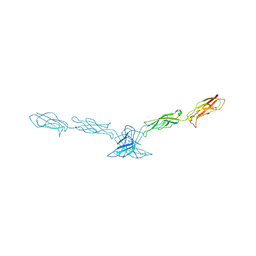 | | Crystal structure of mouse cadherin-8 EC1-3 | | Descriptor: | CALCIUM ION, Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-07-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
6CG6
 
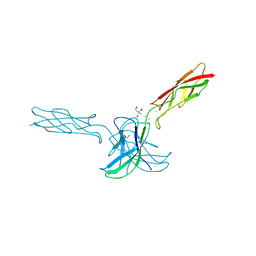 | | mouse cadherin-10 EC1-2 adhesive fragment | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-10, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6CGU
 
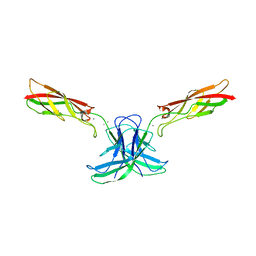 | | mouse cadherin-6 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-6 | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
3LY8
 
 | |
6CG7
 
 | | mouse cadherin-22 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-22 | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6CGS
 
 | | mouse cadherin-7 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-7, GLYCEROL | | Authors: | Brasch, J, Harrison, O.J, Kaczynska, A, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
3LYA
 
 | |
3LY9
 
 | |
