8Q7N
 
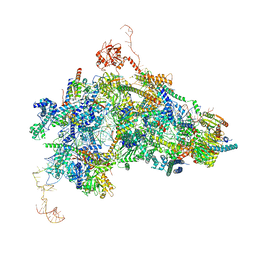 | | cryo-EM structure of the human spliceosomal B complex protomer (tri-snRNP core region) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | New insights into the functions of B complex proteins revealed by cryo-EM of dimerized spliceosomes
To Be Published
|
|
2IO2
 
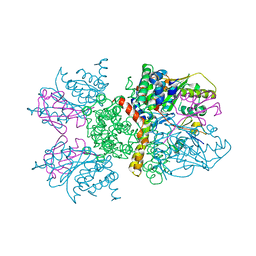 | | Crystal structure of human Senp2 in complex with RanGAP1-SUMO-1 | | Descriptor: | Ran GTPase-activating protein 1, Sentrin-specific protease 2, Small ubiquitin-related modifier 1 | | Authors: | Reverter, D, Lima, C.D. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.
Nat.Struct.Mol.Biol., 13, 2006
|
|
7F60
 
 | |
7F90
 
 | |
7FBV
 
 | |
2J0U
 
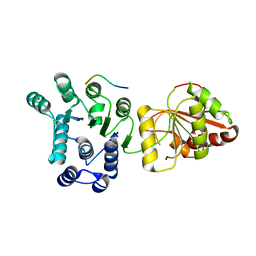 | | The crystal structure of eIF4AIII-Barentsz complex at 3.0 A resolution | | Descriptor: | ATP-DEPENDENT RNA HELICASE DDX48, PROTEIN CASC3 | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
7R5K
 
 | | Human nuclear pore complex (constricted) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science, 376, 2022
|
|
7R5J
 
 | | Human nuclear pore complex (dilated) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-09-21 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (50 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores
Science, 376, 2022
|
|
8QZS
 
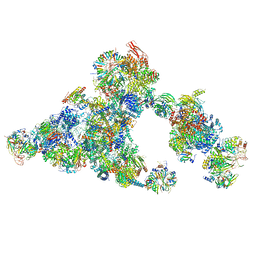 | | Cryo-EM structure of the cross-exon B-like complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Microfibrillar-associated protein 1, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 2024
|
|
8Y0W
 
 | |
6J50
 
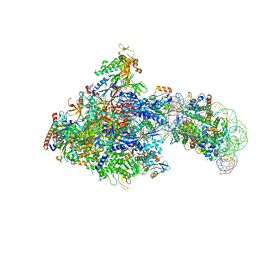 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome (tilted conformation) | | Descriptor: | DNA (198-MER), DNA (41-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4W
 
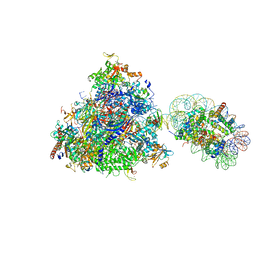 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6JFB
 
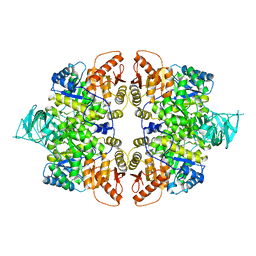 | |
6J4Z
 
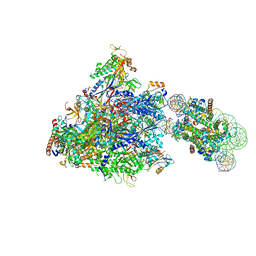 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4X
 
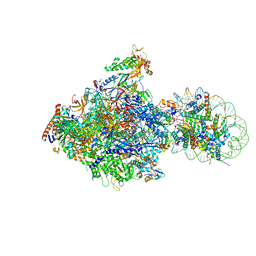 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-1) of the nucleosome (+1A) | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6JOU
 
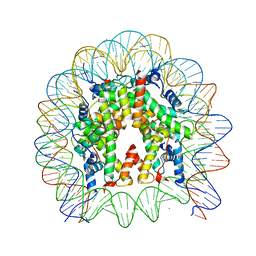 | | Crystal structure of the human nucleosome containing H2A.Z.1 S42R | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Mizukami, Y, Kurumizaka, H. | | Deposit date: | 2019-03-23 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-based design of an H2A.Z.1 mutant stabilizing a nucleosome in vitro and in vivo.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6H3A
 
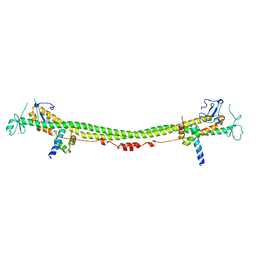 | | Crystal structure of the KAP1 RBCC domain in complex with the SMARCAD1 CUE1 domain. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1, Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Lim, M, Williams, H.L, Svejstrup, J.Q, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-07-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (5.505 Å) | | Cite: | A Ubiquitin-Binding Domain that Binds a Structural Fold Distinct from that of Ubiquitin.
Structure, 27, 2019
|
|
6FF4
 
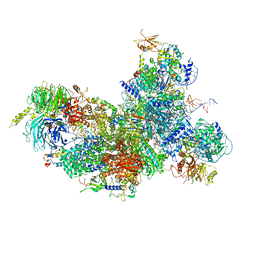 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
6J4I
 
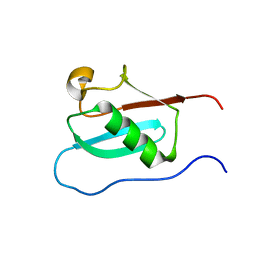 | |
6JXV
 
 | | SUMO1 bound to phosphorylated SLS4-SIM peptide from ICP0 | | Descriptor: | Phosphorylated SLS4-SIM from ubiquitin E3 ligase ICP0, Small ubiquitin-related modifier | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6JXU
 
 | | SUMO1 bound to SLS4-SIM peptide from ICP0 | | Descriptor: | Small ubiquitin-related modifier, viral protein | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
2N1V
 
 | |
2N1A
 
 | | Docked structure between SUMO1 and ZZ-domain from CBP | | Descriptor: | CREB-binding protein, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Diehl, C. | | Deposit date: | 2015-03-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of a Complex between Small Ubiquitin-like Modifier 1 (SUMO1) and the ZZ Domain of CREB-binding Protein (CBP/p300) Reveals a New Interaction Surface on SUMO.
J.Biol.Chem., 291, 2016
|
|
2MW5
 
 | |
2KQS
 
 | |
