7DW8
 
 | | Structure of a novel beta-mannanase BaMan113A with mannobiose, N236Y mutation. | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-15 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
3HRG
 
 | |
3HM4
 
 | |
7LD3
 
 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
2OEJ
 
 | | Crystal structure of a rubisco-like protein from Geobacillus kaustophilus (tetramutant form), liganded with phosphate ions | | Descriptor: | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase, PHOSPHATE ION | | Authors: | Fedorov, A.A, Imker, H.J, Fedorov, E.V, Almo, S.C, Gerlt, J.A. | | Deposit date: | 2006-12-30 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanistic Diversity in the RuBisCO Superfamily: The "Enolase" in the Methionine Salvage Pathway in Geobacillus kaustophilus.
Biochemistry, 46, 2007
|
|
4H57
 
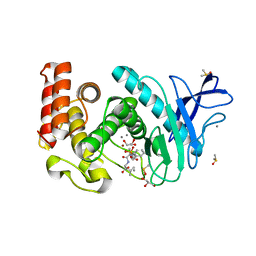 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the hydrophobic effect on the molecular level: the role of water, enthalpy, and entropy in ligand binding to thermolysin.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3HX8
 
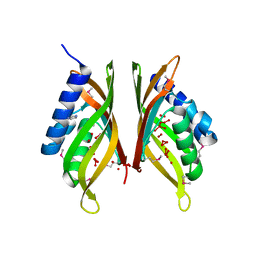 | |
4D5G
 
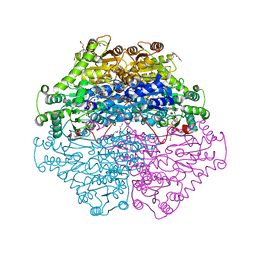 | | Structure of recombinant CDH-H28AN484A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CYCLOHEXANE-1,2-DIONE HYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loschonsky, S, Wacker, T, Waltzer, S, Giovannini, P.P, McLeish, M.J, Andrade, S.L.A, Mueller, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extended Reaction Scope of Thiamine Diphosphate Dependent Cyclohexane-1,2-Dione Hydrolase: From C-C Bond Cleavage to C-C Bond Ligation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CVD
 
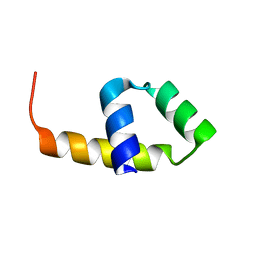 | |
4AR7
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2012-04-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structure of a Fluorescent Protein from Aequorea Victoria Bearing the Obligate-Monomer Mutation A206K.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3KL7
 
 | |
4NW4
 
 | |
1HCZ
 
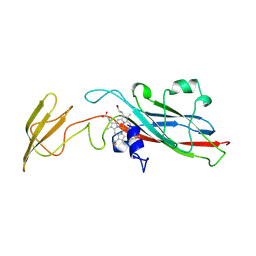 | | LUMEN-SIDE DOMAIN OF REDUCED CYTOCHROME F AT-35 DEGREES CELSIUS | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain.
Protein Sci., 5, 1996
|
|
3HOI
 
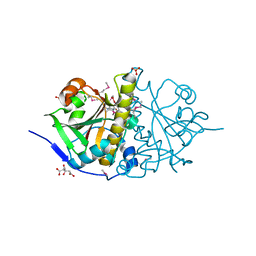 | |
4EB3
 
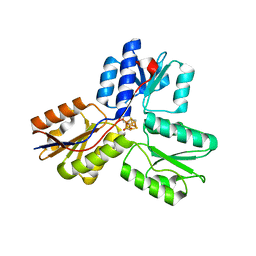 | | Crystal structure of IspH in complex with iso-HMBPP | | Descriptor: | 3-(hydroxymethyl)but-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Wang, W, Wang, K, Span, I, Bacher, A, Groll, M, Oldfield, E. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Are free radicals involved in IspH catalysis? An EPR and crystallographic investigation.
J.Am.Chem.Soc., 134, 2012
|
|
3I0Y
 
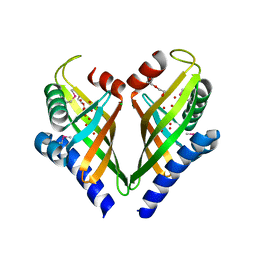 | |
6DA9
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
2Q83
 
 | |
4RGL
 
 | |
3L2N
 
 | |
4LQX
 
 | |
2PY6
 
 | |
5K09
 
 | | Crystal Structure of COMT in complex with a thiazole ligand | | Descriptor: | 5-{3-[(4-methoxyphenyl)methyl]-1H-pyrazol-5-yl}-2,4-dimethyl-1,3-thiazole, Catechol O-methyltransferase, PHOSPHATE ION, ... | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
3H51
 
 | |
3GPT
 
 | | Crystal structure of the yeast 20S proteasome in complex with Salinosporamide derivatives: slow substrate ligand | | Descriptor: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-4-(2-fluoroethyl)-3-hydroxy-3-methyl-5-oxopyrrolidine-2-carbaldehyde, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Macherla, V.R, Manam, R.R, Arthur, K.A.M, Potts, C.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
