3Q1E
 
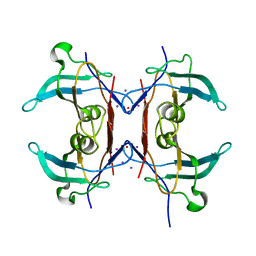 | | Crystal structure of Y116T/I16A double mutant of 5-hydroxyisourate hydrolase in complex with T4 | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Percudani, R, Zanotti, G, Berni, R. | | Deposit date: | 2010-12-17 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
3GR9
 
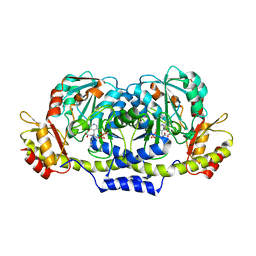 | | Crystal structure of ColD H188K S187N | | Descriptor: | 2-OXOGLUTARIC ACID, ColD | | Authors: | Holden, H.M, Cook, P.D, Kubiak, R.L, Toomey, D.P. | | Deposit date: | 2009-03-25 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Site-Directed Mutations Are Required for the Conversion of a Sugar Dehydratase into an Aminotransferase.
Biochemistry, 48, 2009
|
|
3GST
 
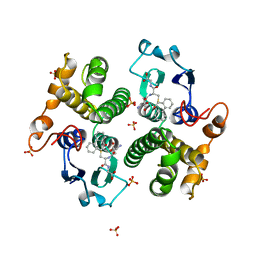 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
4F5I
 
 | |
3GR4
 
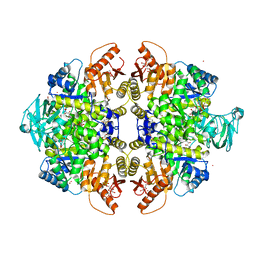 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-[(2,6-difluorophenyl)sulfonyl]-4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)piperazine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
3GYX
 
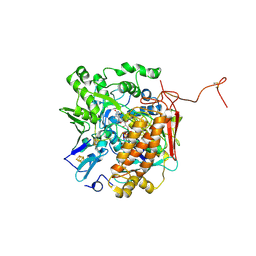 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
3H32
 
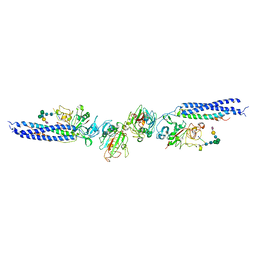 | |
4F1I
 
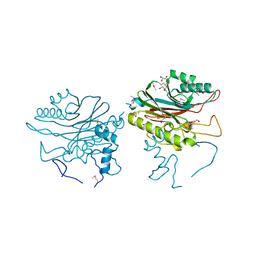 | |
3IWU
 
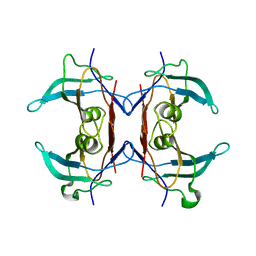 | | Crystal structure of Y116T/I16A double mutant of 5-hydroxyisourate hydrolase | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
3QAS
 
 | | Structure of Undecaprenyl Diphosphate synthase | | Descriptor: | Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Oldfield, E. | | Deposit date: | 2011-01-11 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Applying Molecular Dynamics Simulations to Identify Rarely Sampled Ligand-bound Conformational States of Undecaprenyl Pyrophosphate Synthase, an Antibacterial Target.
Chem.Biol.Drug Des., 77, 2011
|
|
3QBI
 
 | |
3QAK
 
 | | Agonist bound structure of the human adenosine A2a receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,lysozyme chimera | | Authors: | Xu, F, Wu, H, Katritch, V, Han, G.W, Cherezov, V, Stevens, R, GPCR Network (GPCR) | | Deposit date: | 2011-01-11 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an agonist-bound human A2A adenosine receptor.
Science, 332, 2011
|
|
3GQY
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-4-[(4-methoxyphenyl)sulfonyl]piperazine, L(+)-TARTARIC ACID, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
4C7R
 
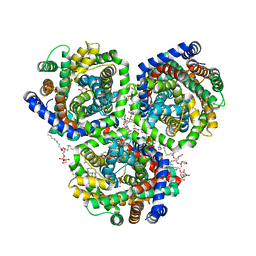 | | Inward facing conformation of the trimeric betaine transporter BetP in complex with lipids | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, CHLORIDE ION, ... | | Authors: | Koshy, C, Yildiz, O, Ziegler, C. | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Evidence for Functional Lipid Interactions in the Betaine Transporter Betp
Embo J., 32, 2013
|
|
3IE3
 
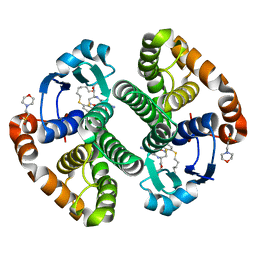 | | Structural basis for the binding of the anti-cancer compound 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) to human glutathione S-transferases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
4C9X
 
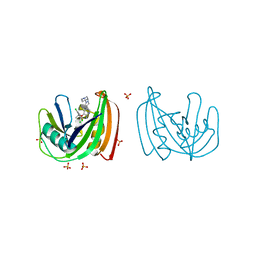 | | Crystal structure of NUDT1 (MTH1) with S-crizotinib | | Descriptor: | 3-[(1S)-1-(2,6-DICHLORO-3-FLUOROPHENYL)ETHOXY]-5-(1-PIPERIDIN-4-YLPYRAZOL-4-YL)PYRIDIN-2-AMINE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
3KZC
 
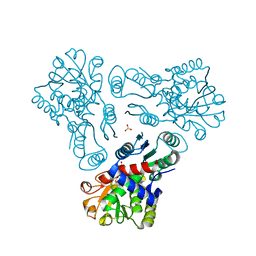 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase | | Descriptor: | N-acetylornithine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.
J.Biol.Chem., 280, 2005
|
|
3GTE
 
 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Iron | | Descriptor: | ACETATE ION, DdmC, FE (III) ION, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
3KZM
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3QJD
 
 | |
3QJV
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3GUS
 
 | | Crystal strcture of human Pi class glutathione S-transferase GSTP1-1 in complex with 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
3KZO
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate and N-acetyl-L-norvaline | | Descriptor: | GLYCEROL, N-ACETYL-L-NORVALINE, N-acetylornithine carbamoyltransferase, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
4C52
 
 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (39b) | | Descriptor: | (R)-2-(3-(3-((2,4-DIFLUOROPENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)-3-(ISOBUTYLTHIO) PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
3R43
 
 | | AKR1C3 complexed with mefenamic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, Aldo-keto reductase family 1 member C3, ... | | Authors: | Squire, C.J, Teague, R.M, Yosaatmadja, L. | | Deposit date: | 2011-03-17 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
