6J5Z
 
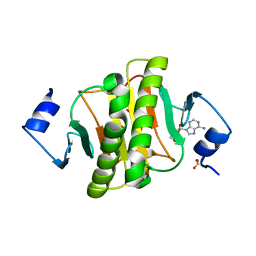 | | Crystal structure of human HINT1 mutant complexing with AP3A | | Descriptor: | ADENOSINE, ETHANESULFONIC ACID, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-12 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6GYE
 
 | | Crystal structure of NadR protein in complex with NR | | Descriptor: | Nicotinamide riboside, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, SULFATE ION | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
6V9A
 
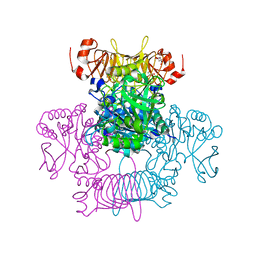 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72D | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6VT9
 
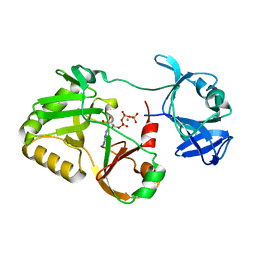 | | Naegleria gruberi RNA ligase E227A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
5TKP
 
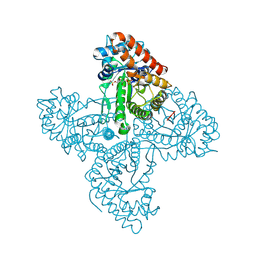 | |
6W10
 
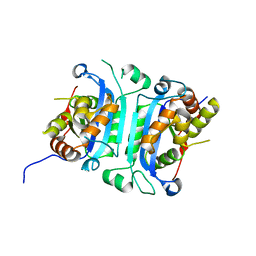 | |
6Q85
 
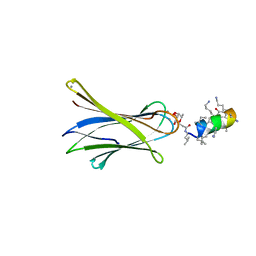 | | Structure of Fucosylated D-antimicrobial peptide SB11 in complex with the Fucose-binding lectin PA-IIL at 1.990 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6Q8H
 
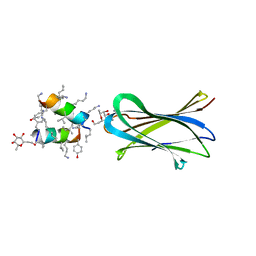 | | Structure of Fucosylated D-antimicrobial peptide SB10 in complex with the Fucose-binding lectin PA-IIL at 1.707 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6VTD
 
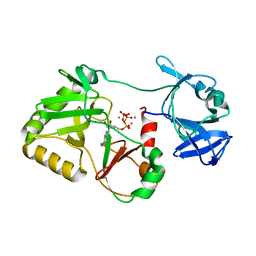 | | Naegleria gruberi RNA ligase R149A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTF
 
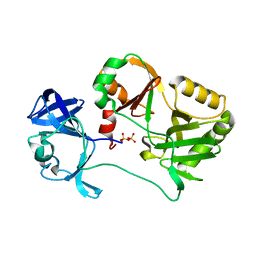 | | Naegleria gruberi RNA ligase with PPi | | Descriptor: | PYROPHOSPHATE 2-, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
5U05
 
 | |
6Q87
 
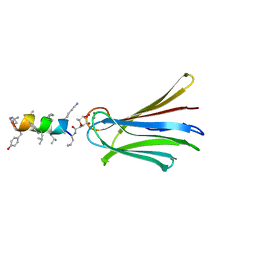 | | Structure of Fucosylated D-antimicrobial peptide SB10 in complex with the Fucose-binding lectin PA-IIL at 2.541 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
5TJS
 
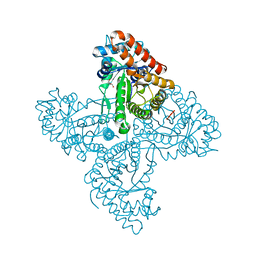 | |
5TK3
 
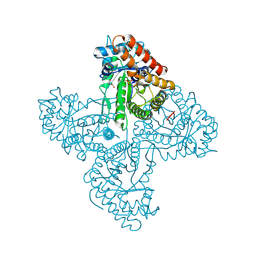 | | Crystal structure of FBP aldolase from Toxoplasma gondii, burst-phase ternary complex | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Heron, P.W, Sygusch, J. | | Deposit date: | 2016-10-06 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Isomer activation controls stereospecificity of class I fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 292, 2017
|
|
5TKN
 
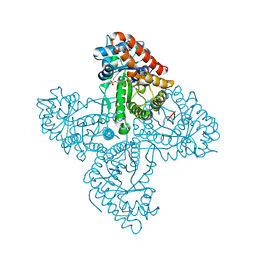 | |
6Q8D
 
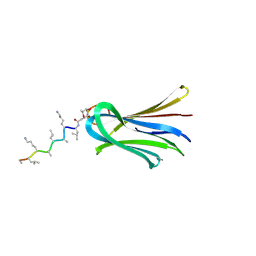 | | Structure of Fucosylated D-antimicrobial peptide SB15 in complex with the Fucose-binding lectin PA-IIL at 1.630 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6QDV
 
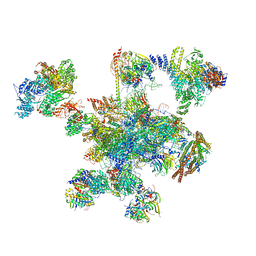 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
6WT9
 
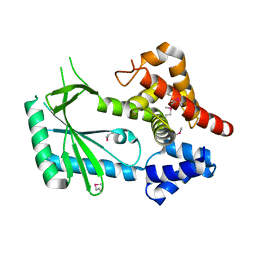 | | Structure of STING-associated CdnE c-di-GMP synthase from Capnocytophaga granulosa | | Descriptor: | NTP_transf_2 domain-containing protein | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6Q6W
 
 | | Structure of Fucosylated D-antimicrobial peptide SB5 in complex with the Fucose-binding lectin PA-IIL at 1.438 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.L. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.438 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6VTB
 
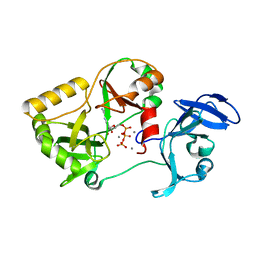 | | Naegleria gruberi RNA ligase K326A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
5SVC
 
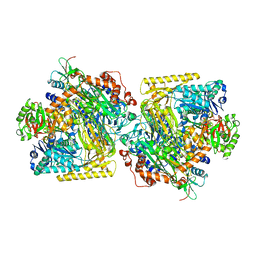 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase nucleotide-free structure | | Descriptor: | Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, Acetone carboxylase gamma subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
6Q86
 
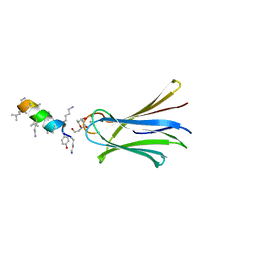 | | Structure of Fucosylated D-antimicrobial peptide SB4 in complex with the Fucose-binding lectin PA-IIL at 2.008 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6Q8G
 
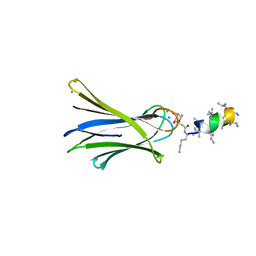 | | Structure of Fucosylated D-antimicrobial peptide SB8 in complex with the Fucose-binding lectin PA-IIL at 1.190 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6Q6X
 
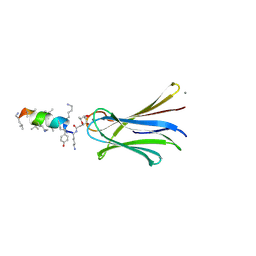 | | Structure of Fucosylated D-antimicrobial peptide SB6 in complex with the Fucose-binding lectin PA-IIL at 1.525 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.525 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6Q77
 
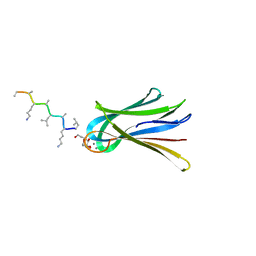 | | Structure of Fucosylated D-antimicrobial peptide SB12 in complex with the Fucose-binding lectin PA-IIL at 2.002 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
