7MHU
 
 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
316D
 
 | | Selectivity of F8-actinomycin D for RNA:DNA hybrids and its anti-leukemia activity | | Descriptor: | 8-FLUORO-ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Takusagawa, F, Takusagawa, K.T, Carlson, R.G, Weaver, R.F. | | Deposit date: | 1997-03-05 | | Release date: | 1997-11-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selectivity of F8-Actinomycin D for RNA:DNA Hybrids and its Anti-Leukemia Activity.
Bioorg.Med.Chem., 5, 1997
|
|
4ARQ
 
 | | Structure of the pesticin S89C, S285C double mutant | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARP
 
 | | Structure of the inactive pesticin E178A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARH
 
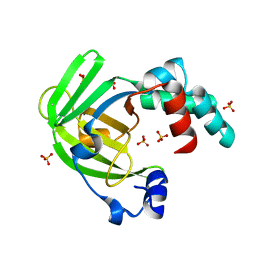 | | X ray structure of the periplasmic zinc binding protein ZinT from Salmonella enterica | | Descriptor: | METAL-BINDING PROTEIN YODA, SULFATE ION | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
6S92
 
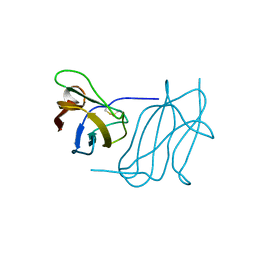 | |
6S94
 
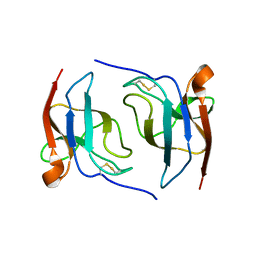 | |
6S95
 
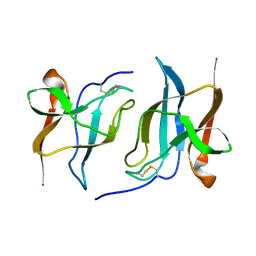 | |
6SBT
 
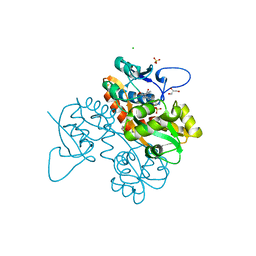 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with N-(7-(1H-imidazol-1-yl)-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl benzamide at 2.3 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Moellerud, S, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2019-07-22 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-(7-(1H-Imidazol-1-yl)-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)benzamide, a New Kainate Receptor Selective Antagonist and Analgesic: Synthesis, X-ray Crystallography, Structure-Affinity Relationships, and in Vitro and in Vivo Pharmacology.
Acs Chem Neurosci, 10, 2019
|
|
6S93
 
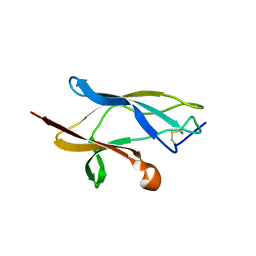 | |
6SNY
 
 | | Synthetic mimic of an EPCR-binding PfEMP1 bound to EPCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Barber, N.M, Higgins, M.K. | | Deposit date: | 2019-08-27 | | Release date: | 2019-09-04 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-Guided Design of a Synthetic Mimic of an Endothelial Protein C Receptor-Binding PfEMP1 Protein.
Msphere, 6, 2021
|
|
4ZJ3
 
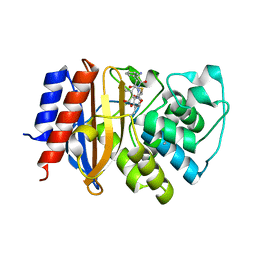 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AIN
 
 | | Varenicline Interactions at the 5HT3 Receptor Ligand Binding Site are Revealed by 5HTBP | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, VARENICLINE | | Authors: | Price, K.L, Lillestol, R, Ulens, C, Lummis, S.C. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Varenicline Interactions at the 5-Ht3 Receptor Ligand Binding Site are Revealed by 5-Htbp.
Acs Chem Neurosci, 6, 2015
|
|
7OWR
 
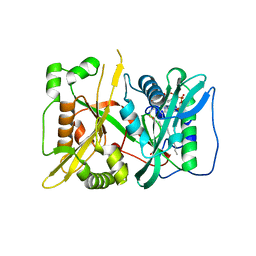 | | HsNMT1 in complex with both MyrCoA and peptide GGKSFSKPR | | Descriptor: | GLY-GLY-LYS-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWQ
 
 | |
7PHP
 
 | | Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, Multidrug resistance protein NorM, NabFab HC, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Niederer, M, Pardon, E, Steyaert, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PIJ
 
 | | Structure of Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMTNb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PHQ
 
 | | Structure of homo-dimeric Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMT-Nb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PDC
 
 | |
7PKX
 
 | | Crystal structure of a DyP-type peroxidase from Bacillus subtilis in P3121 space group | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Borges, P.T, Rodrigues, C, Silva, D, Taborda, A, Brissos, V, Frazao, C, Martins, L.O. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Loops around the Heme Pocket Have a Critical Role in the Function and Stability of Bs DyP from Bacillus subtilis .
Int J Mol Sci, 22, 2021
|
|
6MQD
 
 | | Myotoxin II from Bothrops moojeni complexed with Rosmarinic Acid | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, Basic phospholipase A2 homolog 2 | | Authors: | Salvador, G.H.M, Fontes, M.R.M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Search for efficient inhibitors of myotoxic activity induced by ophidian phospholipase A2-like proteins using functional, structural and bioinformatics approaches.
Sci Rep, 9, 2019
|
|
6MQF
 
 | |
8BC9
 
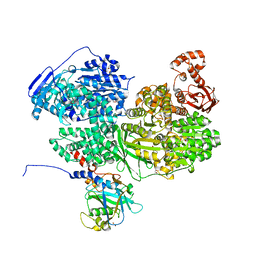 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 24 | | Descriptor: | 1,2-ETHANEDIOL, N-hydroxybenzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCH
 
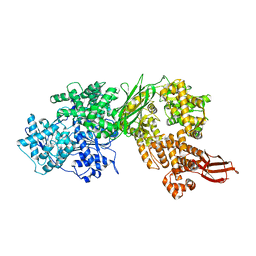 | | Human Brr2 Helicase Region in complex with Sulfaguanidine | | Descriptor: | 1-(4-aminophenyl)sulfonylguanidine, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCC
 
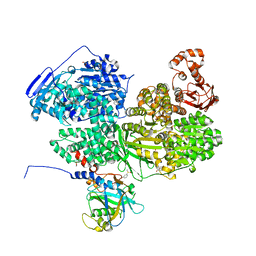 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 39 | | Descriptor: | 1,2-ETHANEDIOL, 3-oxidanylbenzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
