3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
6SP0
 
 | |
5JNM
 
 | | Crystal structure of MtlD from Staphylococcus aureus at 1.7-Angstrom resolution | | Descriptor: | Mannitol-1-phosphate 5-dehydrogenase, SULFATE ION | | Authors: | Ta, H.M, Nguyen, T, Kim, T, Kim, K.K. | | Deposit date: | 2016-04-30 | | Release date: | 2017-11-08 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Targeting Mannitol Metabolism as an Alternative Antimicrobial Strategy Based on the Structure-Function Study of Mannitol-1-Phosphate Dehydrogenase in Staphylococcus aureus.
Mbio, 10, 2019
|
|
7PRW
 
 | | The glucocorticoid receptor in complex with velsecorat, a PGC1a coactivator fragment and sgk 23bp | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*AP*CP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*GP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*CP*GP*GP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*TP*GP*TP*AP*C)-3'), ... | | Authors: | Postel, S, Edman, K, Wissler, L. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PRV
 
 | | The glucocorticoid receptor in complex with fluticasone furoate, a PGC1a coactivator fragment and sgk 23bp | | Descriptor: | (6alpha,11alpha,14beta,16alpha,17alpha)-6,9-difluoro-17-{[(fluoromethyl)sulfanyl]carbonyl}-11-hydroxy-16-methyl-3-oxoan drosta-1,4-dien-17-yl furan-2-carboxylate, 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*GP*TP*CP*GP*A)-3'), ... | | Authors: | Postel, S, Edman, K, Wissler, L. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PRX
 
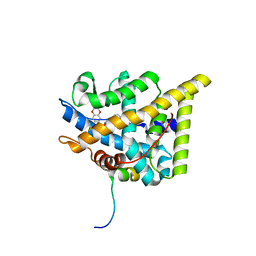 | | wildtype ligand binding domain of the glucocorticoid receptor complexed with velsecorat and a PGC1a coactivator fragment | | Descriptor: | Glucocorticoid receptor, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Velsecorat | | Authors: | Edman, K, Wissler, L, Koehler, C, Postel, S. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4IGS
 
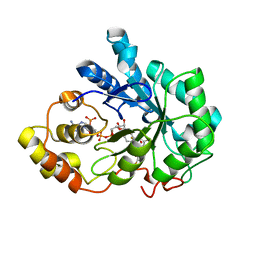 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5J5F
 
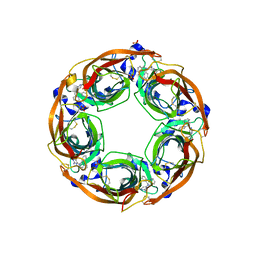 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with N4,N4-bis[(pyridin-2-yl)methyl]-6-(thiophen-3-yl)pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczanowska, K, Camacho Hernandez, G.A, Harel, M, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
2BNI
 
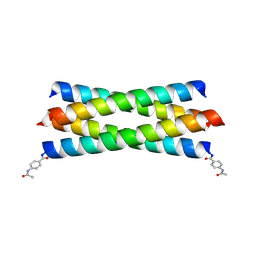 | | pLI mutant E20C L16G Y17H, antiparallel | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2005-03-24 | | Release date: | 2005-03-30 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1PCL
 
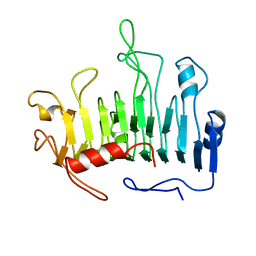 | |
5JNQ
 
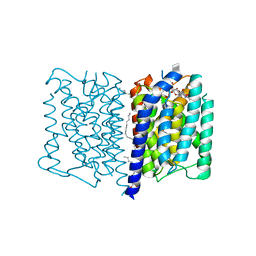 | | MraY tunicamycin complex | | Descriptor: | PALMITIC ACID, Phospho-N-acetylmuramoyl-pentapeptide-transferase, Tunicamycin, ... | | Authors: | Johansson, P. | | Deposit date: | 2016-04-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MraY-antibiotic complex reveals details of tunicamycin mode of action.
Nat. Chem. Biol., 13, 2017
|
|
8AEZ
 
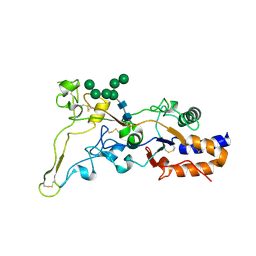 | | X-ray structure of the deglycosylated receptor binding domain of Env glycoprotein of Simian Foamy virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp130, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Backovic, M. | | Deposit date: | 2022-07-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | The crystal structure of a simian Foamy Virus receptor binding domain provides clues about entry into host cells.
Nat Commun, 14, 2023
|
|
5K1I
 
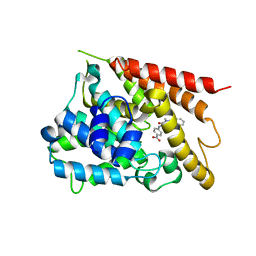 | | PDE4 crystal structure in complex with small molecule inhibitor | | Descriptor: | 4-[(5-acetyl-2-ethyl-3-oxo-6-phenyl-2,3-dihydropyridazin-4-yl)amino]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Segarra, V, Hernandez, B, Ferrer-Miralles, N, Korndoerfer, I, Aymami, J. | | Deposit date: | 2016-05-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Biphenyl Pyridazinone Derivatives as Inhaled PDE4 Inhibitors: Structural Biology and Structure-Activity Relationships.
J. Med. Chem., 59, 2016
|
|
6YBA
 
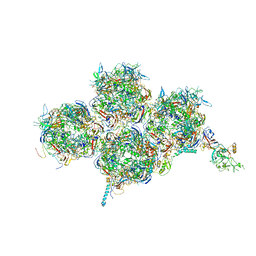 | | HAdV-F41 Capsid | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Perez Illana, M, Martinez, M, Mangroo, C, Brown, M, Marabini, R, San Martin, C. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses.
Sci Adv, 7, 2021
|
|
5JXK
 
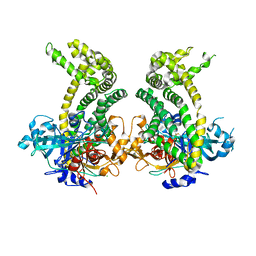 | | Crystal structure of Porphyromonas endodontalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K7F
 
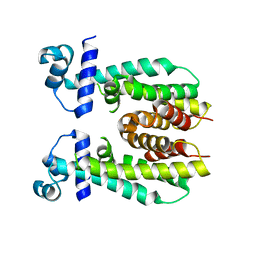 | | Crystal structure of apo AibR | | Descriptor: | ACETATE ION, Transcriptional regulator, TetR family | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
6VOS
 
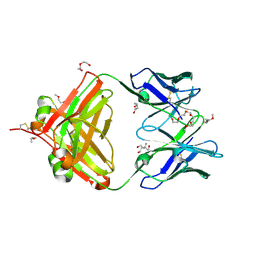 | | Crystal structure of macaque anti-HIV-1 antibody RM20J | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VSR
 
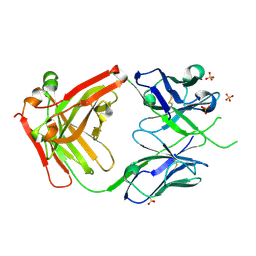 | |
6YQY
 
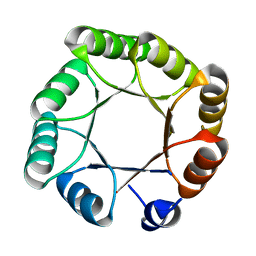 | | Crystal structure of sTIM11noCys, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel sTIM11noCys | | Authors: | Romero-Romero, S, Wiese, G.J, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6YQX
 
 | | Crystal structure of DeNovoTIM13, a de novo designed TIM barrel | | Descriptor: | CHLORIDE ION, GLYCEROL, de novo designed TIM barrel DeNovoTIM13 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
2C4I
 
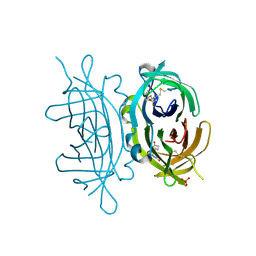 | | Crystal structure of engineered avidin | | Descriptor: | AVIDIN, BIOTIN, SULFATE ION | | Authors: | Hytonen, V.P, Horha, J, Airenne, T.T, Niskanen, E.A, Helttunen, K, Johnson, M.S, Salminen, T.A, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Controlling Quaternary Structure Assembly: Subunit Interface Engineering and Crystal Structure of Dual Chain Avidin.
J.Mol.Biol., 359, 2006
|
|
6VN0
 
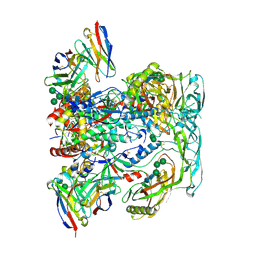 | | BG505 SOSIP.v4.1 in complex with rhesus macaque Fab RM20F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Cottrell, C.A, Shin, M, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-24 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VLR
 
 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 and PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIPv5.2 gp41, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-24 | | Release date: | 2020-06-24 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
5JY0
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with substrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Asp/Glu-specific dipeptidyl-peptidase, LEU-ASP-VAL | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K32
 
 | | PDE4D crystal structure in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-methoxyphenyl)amino]-2-phenyl-7,8-dihydro-1,6-naphthyridin-5(6H)-one, MAGNESIUM ION, ... | | Authors: | Segarra, V, Hernandez, B, Roberts, R, Gracia, J, Soler, M, Bonin, I, Aymami, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 4-Amino-7,8-dihydro-1,6-naphthyridin-5(6 H)-ones as Inhaled Phosphodiesterase Type 4 (PDE4) Inhibitors: Structural Biology and Structure-Activity Relationships.
J. Med. Chem., 61, 2018
|
|
