6NVE
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 7 | | Descriptor: | 4-{[(1S,2S)-2-(carboxymethyl)cyclopentyl]methyl}benzoic acid, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 7
To Be Published
|
|
6VSG
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 4-(trifluoromethyl)benzene-1,2-diamine(fragment 17) | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VU5
 
 | | Structure of G-alpha-q bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(q) subunit alpha, Resistance to inhibitors of cholinesterase-8A (Ric-8A) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
5LLV
 
 | | Crystal structure of DACM F87M/L110M Transthyretin mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, Transthyretin | | Authors: | Sala, B.M, Ghadami, S.A, Bemporad, F, Chiti, F, Ricagno, S. | | Deposit date: | 2016-07-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | FRET studies of various conformational states adopted by transthyretin.
Cell. Mol. Life Sci., 74, 2017
|
|
7CLV
 
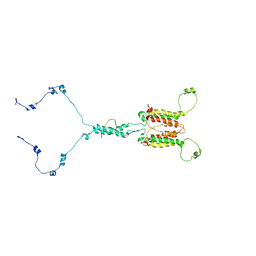 | | Solution structure of mitochondrial Tim23 channel in complex with a signaling peptide | | Descriptor: | COX4 isoform 1, TIM23 isoform 1 | | Authors: | Zhou, S, Ruan, M.S, Li, Y.Y, Yang, J, Richter, C, Schwalbe, H, Shen, B, Wang, J.F. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide.
Cell Res., 31, 2021
|
|
6NXE
 
 | |
1MCK
 
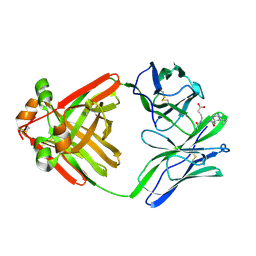 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-D-GLU-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
7LZE
 
 | | Cryo-EM Structure of disulfide stabilized HMPV F v4-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6VMY
 
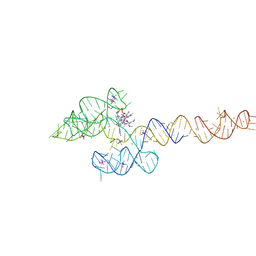 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
8BWC
 
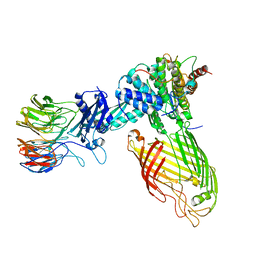 | | E. coli BAM complex (BamABCDE) wild-type | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Machin, J.M, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-12-06 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Darobactin B Stabilises a Lateral-Closed Conformation of the BAM Complex in E. coli Cells.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1MCQ
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
7LDW
 
 | |
7LD2
 
 | |
4QPZ
 
 | | Crystal structure of the formolase FLS_v2 in space group P 21 | | Descriptor: | Formolase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L, Baker, D. | | Deposit date: | 2014-06-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5EIP
 
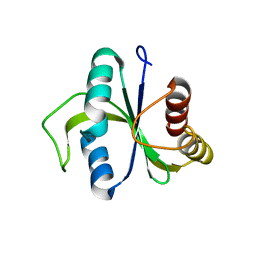 | | apo-structure of YTH domain of SpMmi1 | | Descriptor: | YTH domain-containing protein mmi1 | | Authors: | Wu, B.X, Xu, J.H, Su, S.C, Ma, J.B. | | Deposit date: | 2015-10-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1
Biochem. Biophys. Res. Commun., 491, 2017
|
|
1AEJ
 
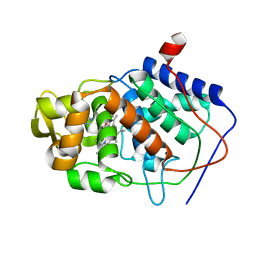 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (1-VINYLIMIDAZOLE) | | Descriptor: | 1-VINYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
7LBZ
 
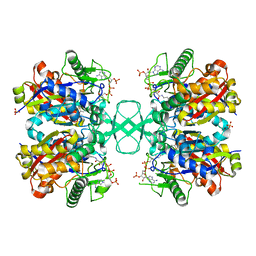 | |
5UUN
 
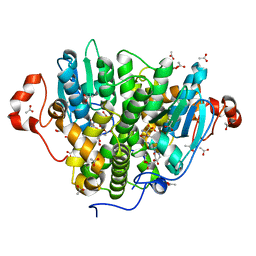 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
5LKK
 
 | |
8BV1
 
 | | Peptide inhibitor P4 in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, P4 peptide inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.834 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
5K5W
 
 | | Crystal structure of limiting CO2-inducible protein LCIB | | Descriptor: | ZINC ION, limiting CO2-inducible protein LCIB | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Cajar, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5MDQ
 
 | |
460D
 
 | |
1AEH
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (2-AMINO-4-METHYLTHIAZOLE) | | Descriptor: | 2-AMINO-4-METHYLTHIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
