5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
7UBZ
 
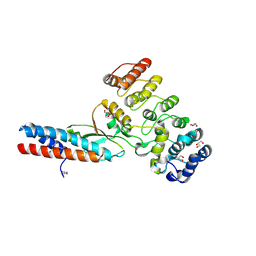 | | Chymotrypsin digested toxin/immunity complex for a T6SS lipase effector from E. cloacae | | Descriptor: | Ankyrin repeat domain-containing protein, CITRATE ANION, GLYCEROL, ... | | Authors: | Cuthbert, B.J, Jensen, S.J, Goulding, C.W, Hayes, C.S. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Toxin/immunity complex for a T6SS lipase effector from E. cloacae
To Be Published
|
|
4Q8G
 
 | |
6VSJ
 
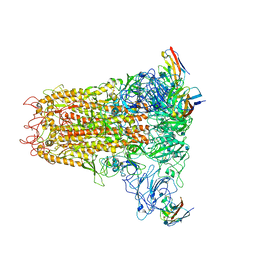 | | Cryo-electron microscopy structure of mouse coronavirus spike protein complexed with its murine receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, Spike glycoprotein | | Authors: | Shang, J, Wan, Y.S, Liu, C, Yount, B, Gully, K, Yang, Y, Auerbach, A, Peng, G.Q, Baric, R, Li, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry.
Plos Pathog., 16, 2020
|
|
7UT1
 
 | |
5NG1
 
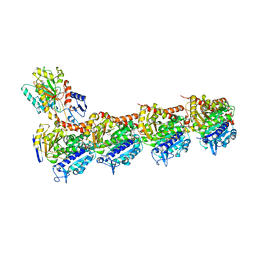 | | TUBULIN-MTC-zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, (2~{Z},4~{E})-~{N}-[(~{S})-oxidanyl-[(1~{S},2~{E},5~{S},11~{R},17~{S},19~{R})-3,11,19-trimethyl-7,13-bis(oxidanylidene)-6,21-dioxabicyclo[15.3.1]henicos-2-en-5-yl]methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
2WLL
 
 | | POTASSIUM CHANNEL FROM BURKHOLDERIA PSEUDOMALLEI | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, MAGNESIUM ION, POTASSIUM CHANNEL, ... | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, VandenBerg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
1BDV
 
 | | ARC FV10 COCRYSTAL | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (ARC FV10 REPRESSOR) | | Authors: | Schildbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5K5T
 
 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
6HQF
 
 | | Structure of Phenylalanine ammonia-lyase from Petroselinum crispum in complex with (R)-APEP | | Descriptor: | Phenylalanine ammonia-lyase 1, [(1R)-1-amino-2-phenylethyl]phosphonic acid | | Authors: | Bata, Z, Molnar, B, Leveles, I, Poppe, L, Vertessy, G.B. | | Deposit date: | 2018-09-24 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Substrate Tunnel Engineering Aided by X-ray Crystallography and Functional Dynamics Swaps the Function of MIO-Enzymes
Acs Catalysis, 2021
|
|
4YUN
 
 | | Multiconformer synchrotron model of CypA at 310 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
5K8C
 
 | | X-ray structure of KdnB, 3-deoxy-alpha-D-manno-octulosonate 8-oxidase, from Shewanella oneidensis | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-alpha-D-manno-octulosonate 8-oxidase, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B, Zachman-Brockmeyer, T.R. | | Deposit date: | 2016-05-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of KdnB and KdnA from Shewanella oneidensis: Key Enzymes in the Formation of 8-Amino-3,8-Dideoxy-d-Manno-Octulosonic Acid.
Biochemistry, 55, 2016
|
|
2OIY
 
 | |
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
6BH8
 
 | | Crystal structure of ZMPSTE24 in complex with phosphoramidon | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CAAX prenyl protease 1 homolog, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Goblirsch, B.R, Arachea, B.T, Wiener, M.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Phosphoramidon inhibits the integral membrane protein zinc metalloprotease ZMPSTE24.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5MLA
 
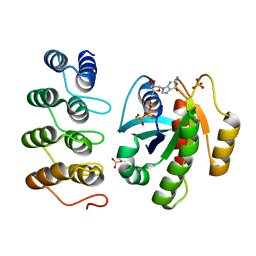 | | Crystal structure of human RAS in complex with darpin K55 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Inhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
7UEB
 
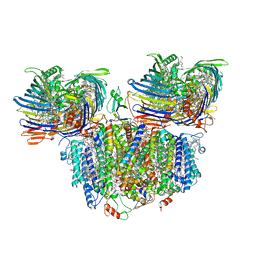 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO2) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
6FKS
 
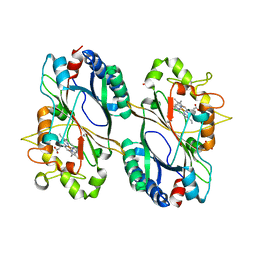 | | Crystal structure of a dye-decolorizing peroxidase from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-24 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.60000467 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
7BCJ
 
 | |
7UK1
 
 | |
5K5P
 
 | |
7UEA
 
 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
5T4J
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, HTH-type transcriptional regulatory protein GabR, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7FQJ
 
 | | Crystal Structure of human Legumain in complex with (2S)-N-[(1S)-3-amino-1-cyano-3-oxopropyl]-1-[1-[4-[(2,4-difluorophenyl)methoxy]phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
6NMN
 
 | |
