3KRD
 
 | |
1Z0N
 
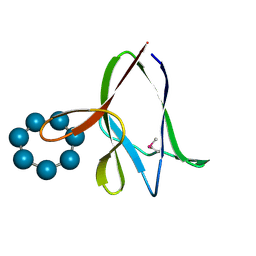 | | the glycogen-binding domain of the AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase, beta-1 subunit, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Polekhina, G, Gupta, A, van Denderen, B.J, Feil, S.C, Kemp, B.E, Stapleton, D, Parker, M.W. | | Deposit date: | 2005-03-02 | | Release date: | 2005-10-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Basis for Glycogen Recognition by AMP-Activated Protein Kinase.
Structure, 13, 2005
|
|
1QWG
 
 | | Crystal structure of Methanococcus jannaschii phosphosulfolactate synthase | | Descriptor: | (2R)-phospho-3-sulfolactate synthase, SULFATE ION | | Authors: | Wise, E.L, Graham, D.E, White, R.H, Rayment, I. | | Deposit date: | 2003-09-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural determination of phosphosulfolactate synthase from Methanococcus jannaschii at 1.7-A resolution: an enolase that is not an enolase
J.Biol.Chem., 278, 2003
|
|
1QG3
 
 | |
1Z1A
 
 | |
6ZLG
 
 | | Folding of an iron binding peptide in response to sedimentation is resolved using ferritin as a nano-reactor | | Descriptor: | Ferritin | | Authors: | Davidov, G, Abelya, G, Zalk, R, Izbicki, B, Shaibi, S, Spektor, L, Meyron Holtz, E.G, Zarivach, R, Frank, G.A. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Folding of an Intrinsically Disordered Iron-Binding Peptide in Response to Sedimentation Revealed by Cryo-EM.
J.Am.Chem.Soc., 142, 2020
|
|
2DEA
 
 | |
1QZ5
 
 | | Structure of rabbit actin in complex with kabiramide C | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Klenchin, V.A, Allingham, J.S, King, R, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin
Nat.Struct.Biol., 10, 2003
|
|
1Q7Y
 
 | | Crystal Structure of CCdAP-Puromycin bound at the Peptidyl transferase center of the 50S ribosomal subunit | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights Into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3KX3
 
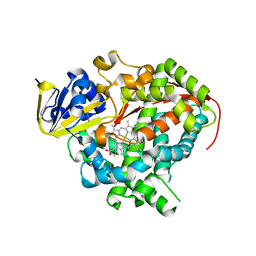 | | Crystal structure of Bacillus megaterium BM3 heme domain mutant L86E | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Girvan, H.M, Levy, C.W, Leys, D, Munro, A.W. | | Deposit date: | 2009-12-02 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Glutamate-haem ester bond formation is disfavoured in flavocytochrome P450 BM3: characterization of glutamate substitution mutants at the haem site of P450 BM3.
Biochem.J., 427, 2010
|
|
3KZ4
 
 | | Crystal Structure of the Rotavirus Double Layered Particle | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Mcclain, B, Settembre, E.C, Bellamy, A.R, Harrison, S.C. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution.
J.Mol.Biol., 397, 2010
|
|
1ZF4
 
 | | ATC Four-stranded DNA Holliday Junction | | Descriptor: | 5'-D(*CP*CP*GP*AP*TP*AP*TP*CP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFF
 
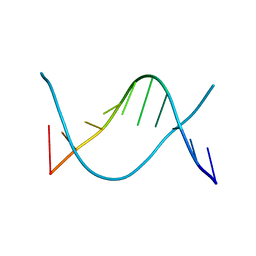 | | TTC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*TP*TP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZGL
 
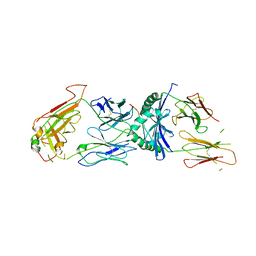 | | Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, Myelin basic protein, ... | | Authors: | Li, Y, Huang, Y, Lue, J, Quandt, J.A, Martin, R, Mariuzza, R.A. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-18 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
Embo J., 24, 2005
|
|
3KIP
 
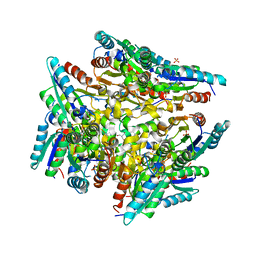 | | Crystal structure of type-II 3-dehydroquinase from C. albicans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinase, type II, ... | | Authors: | Trapani, S, Schoehn, G, Navaza, J, Abergel, C. | | Deposit date: | 2009-11-02 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2DUI
 
 | |
2DUT
 
 | |
1QO1
 
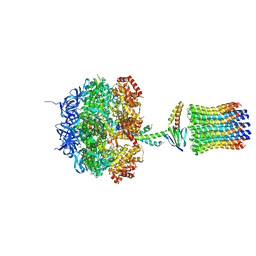 | | Molecular Architecture of the Rotary Motor in ATP Synthase from Yeast Mitochondria | | Descriptor: | ATP SYNTHASE ALPHA CHAIN, ATP SYNTHASE BETA CHAIN, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Stock, D, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1999-11-01 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture of the Rotary Motor in ATP Synthase
Science, 286, 1999
|
|
3KM4
 
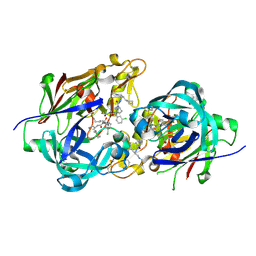 | | Optimization of Orally Bioavailable Alkyl Amine Renin Inhibitors | | Descriptor: | (3R)-3-[(1S)-4-(acetylamino)-1-(3-chlorophenyl)-1-hydroxybutyl]-N-{(1S)-2-cyclohexyl-1-[(methylamino)methyl]ethyl}piperidine-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of orally bioavailable alkyl amine renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1QPB
 
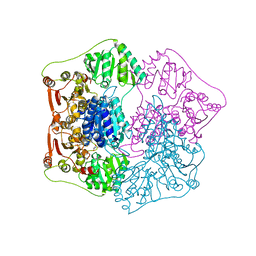 | | PYRUVATE DECARBOYXLASE FROM YEAST (FORM B) COMPLEXED WITH PYRUVAMIDE | | Descriptor: | MAGNESIUM ION, PYRUVAMIDE, PYRUVATE DECARBOXYLASE (FORM B), ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1999-11-26 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis of Substrate Activation in Yeast Pyruvate Decarboxylase a Crystallographic and Kinetic Study
Eur.J.Biochem., 267, 2000
|
|
3KQ7
 
 | |
3KTM
 
 | | Structure of the Heparin-induced E1-Dimer of the Amyloid Precursor Protein (APP) | | Descriptor: | (3R)-butane-1,3-diol, ACETATE ION, Amyloid beta A4 protein, ... | | Authors: | Dahms, S.O, Hoefgen, S, Roeser, D, Schlott, B, Guhrs, K.H, Than, M.E. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ACP
 
 | | Crystal structure of nitrophorin 2 aqua complex | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-07-19 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
1QVG
 
 | | Structure of CCA oligonucleotide bound to the tRNA binding sites of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
1Q86
 
 | | Crystal structure of CCA-Phe-cap-biotin bound simultaneously at half occupancy to both the A-site and P-site of the the 50S ribosomal Subunit. | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into peptide bond formation.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
