7DDT
 
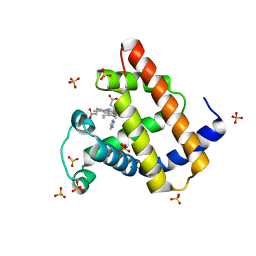 | | Ancestral myoglobin aMbSe of Enaliarctos relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSe, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
3WOG
 
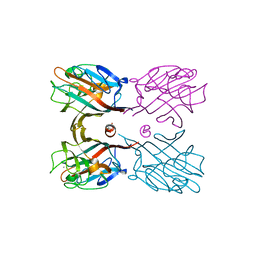 | | Crystal structure plant lectin in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-12-26 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
7DDR
 
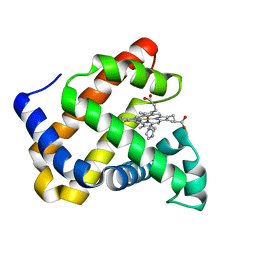 | | Ancestral myoglobin aMbSp of Puijila Darwini relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
2EGK
 
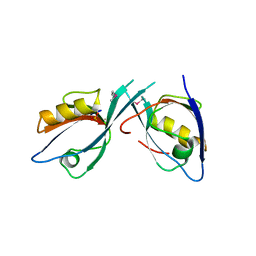 | | Crystal Structure of Tamalin PDZ-Intrinsic Ligand Fusion Protein | | Descriptor: | General receptor for phosphoinositides 1-associated scaffold protein, PHOSPHATE ION | | Authors: | Sugi, T, Oyama, T, Muto, T, Nakanishi, S, Morikawa, K, Jingami, H. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of autoinhibitory PDZ domain of Tamalin: implications for metabotropic glutamate receptor trafficking regulation
Embo J., 26, 2007
|
|
1ZWH
 
 | |
2EAT
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA and TG | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6Y4B
 
 | |
6EW6
 
 | | Crystal structure of the BCL6 BTB domain in complex with anilinopyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}2-(2-chlorophenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC To Provide Insight into Small Molecule Targeting of BCL6.
ACS Chem. Biol., 13, 2018
|
|
1DG6
 
 | | CRYSTAL STRUCTURE OF APO2L/TRAIL | | Descriptor: | APO2L/TNF-RELATED APOPOTIS INDUCING LIGAND (TRAIL), CHLORIDE ION, ZINC ION | | Authors: | Hymowitz, S.G, O'ConnelL, M.P, Ultsch, M.H, de Vos, A.M, Kelley, R.F. | | Deposit date: | 1999-11-23 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A unique zinc-binding site revealed by a high-resolution X-ray structure of homotrimeric Apo2L/TRAIL.
Biochemistry, 39, 2000
|
|
6Y42
 
 | |
3R7D
 
 | | Crystal Structure of Unliganded Aspartate Transcarbamoylase from Bacillus subtilis | | Descriptor: | Aspartate carbamoyltransferase, PHOSPHATE ION | | Authors: | Puleo, D.E, Harris, K.M, Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2011-03-22 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Crystallographic Snapshots of the Complete Catalytic Cycle of the Unregulated Aspartate Transcarbamoylase from Bacillus subtilis.
J.Mol.Biol., 411, 2011
|
|
2EH5
 
 | |
6XFO
 
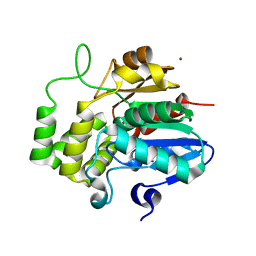 | |
1E3Z
 
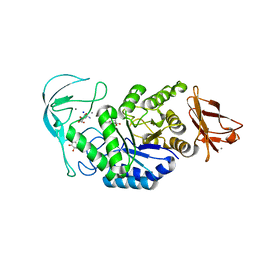 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1H9Y
 
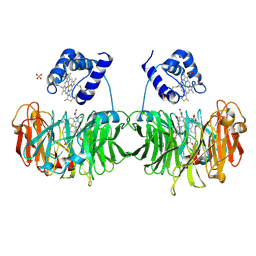 | |
8GN6
 
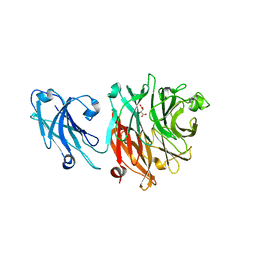 | | Crystallization of Sialidase from Porphyromonas gingivalis | | Descriptor: | Sialidase, UNKNOWN LIGAND | | Authors: | Dong, W.B. | | Deposit date: | 2022-08-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and enzymatic characterization of the sialidase SiaPG from Porphyromonas gingivalis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FDM
 
 | | Human Hemoglobin in Complex with Nitrosomethane | | Descriptor: | GLYCEROL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Powell, S.M, Richter-Addo, G.B, Thomas, L.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structural investigations of heme protein derivatives resulting from reactions of aryl- and alkylhydroxylamines with human hemoglobin.
J.Inorg.Biochem., 246, 2023
|
|
1E43
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1LY2
 
 | | Crystal structure of unliganded human CD21 SCR1-SCR2 (Complement receptor type 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, complement receptor type 2 | | Authors: | Prota, A.E, Sage, D.R, Stehle, T, Fingeroth, J.D. | | Deposit date: | 2002-06-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of human CD21: Implications for Epstein-Barr virus and C3d binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6Y45
 
 | | Crystal Structure of the H33A variant of RsrR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rohac, R, Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Electron and Proton Transfers Modulate DNA Binding by the Transcription Regulator RsrR.
J.Am.Chem.Soc., 142, 2020
|
|
1H9X
 
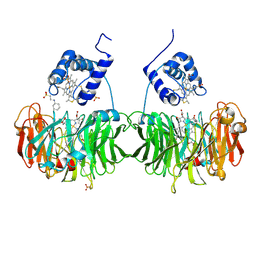 | | Cytochrome cd1 Nitrite Reductase, reduced form | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYTOCHROME CD1 NITRITE REDUCTASE, HEME C, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Alternative Form of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase
J.Biol.Chem., 276, 2001
|
|
6VK3
 
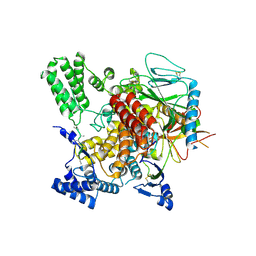 | | CryoEM structure of Hrd3/Yos9 complex | | Descriptor: | Hrd3, Protein OS-9 homolog | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
3ZKI
 
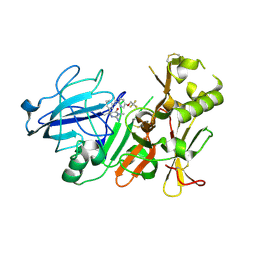 | | BACE2 MUTANT STRUCTURE WITH LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6Y0A
 
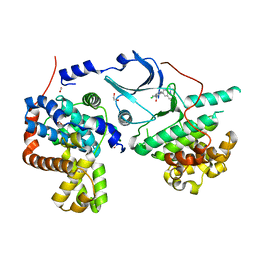 | | CRYSTAL STRUCTURE OF CDK8-CycC IN COMPLEX WITH BI00690300 | | Descriptor: | 1,2-ETHANEDIOL, 6-[5-chloranyl-4-[(1~{S})-1-oxidanylethyl]pyridin-3-yl]-3,4-dihydro-2~{H}-1,8-naphthyridine-1-carboxamide, Cyclin-C, ... | | Authors: | Bader, G, Boettcher, J. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Human Cyclin-Dependent Kinase8/Cyclinc In Complex With Ligand 30180596
To Be Published
|
|
8FDJ
 
 | | Wild-Type Sperm Whale Myoglobin in Complex with Nitrosobenzene | | Descriptor: | Myoglobin, NITROSOBENZENE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Powell, S.M, Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2022-12-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structural investigations of heme protein derivatives resulting from reactions of aryl- and alkylhydroxylamines with human hemoglobin.
J.Inorg.Biochem., 246, 2023
|
|
