5YKZ
 
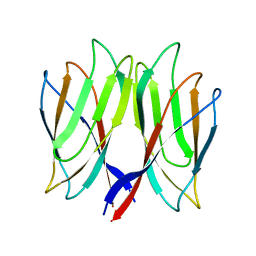 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P21) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
4UBF
 
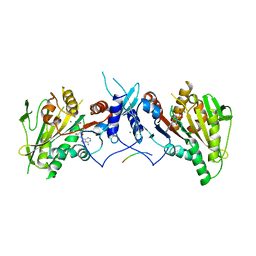 | | HsMCAK motor domain complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION | | Authors: | Welburn, J.P.I, Talapatra, S.K. | | Deposit date: | 2014-08-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-terminal region of the motor protein MCAK controls its structure and activity through a conformational switch.
Elife, 4, 2015
|
|
5YL6
 
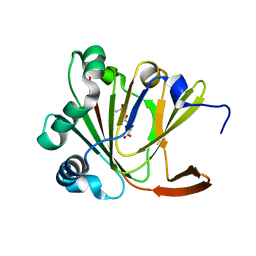 | | Crystal structure of Ofd2 in complex with 2OG from Schizosaccharomyces pombe | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Wang, J, Gao, Y.X, Guo, G.R, Zhu, Z.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2017-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Ofd2 in complex with 2OG from Schizosaccharomyces pombe
to be published
|
|
4UC2
 
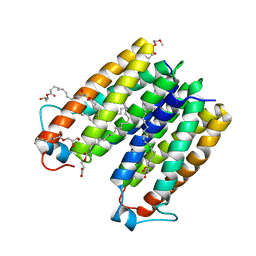 | | Crystal structure of translocator protein 18kDa (TSPO) from rhodobacter sphaeroides (A139T mutant) in P212121 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, TETRAETHYLENE GLYCOL, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
5YLB
 
 | | Crystal structure of Ofd2 from Schizosaccharomyces pombe at 1.80 A | | Descriptor: | FE (III) ION, Uncharacterized protein P8A3.02c | | Authors: | Wang, J, Gao, Y.X, Guo, G.R, Zhu, Z.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of Ofd2 from Schizosaccharomyces pombe at 1.80 A
to be published
|
|
4UMF
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion, Phosphate ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5YMW
 
 | | Crystal structure of 8-mer peptide from Rous sarcoma virus in complex with BF2*1201 | | Descriptor: | Beta-2-microglobulin, Class I histocompatibility antigen, F10 alpha chain, ... | | Authors: | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
4UNR
 
 | | Mtb TMK in complex with compound 23 | | Descriptor: | 4-[3-cyano-2-oxo-7-(1H-pyrazol-4-yl)-5,6-dihydro-1H-benzo[h]quinolin-4-yl]benzoic acid, MAGNESIUM ION, Thymidylate kinase | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
5YNC
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM beta-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4UNY
 
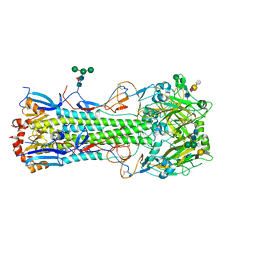 | | Structure of the A_Equine_Newmarket_2_93 H3 haemagglutinin in complex with 6SO4-3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3 HAEMAGGLUTININ HA2 CHAIN, ... | | Authors: | Vachieri, S.G, Collins, P.J, Haire, L.F, Ogrodowicz, R.W, Martin, S.R, Walker, P.A, Xiong, X, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recent Evolution of Equine Influenza and the Origin of Canine Influenza.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KBB
 
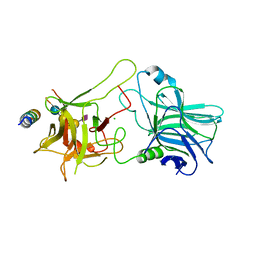 | | Structure of Botulinum neurotoxin B binding domain in complex with both synaptotagmin II and GD1a | | Descriptor: | Botulinum neurotoxin type B, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Berntsson, R.P.A, Peng, L, Dong, M, Stenmark, P. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of dual receptor binding to botulinum neurotoxin B.
Nat Commun, 4, 2013
|
|
5YLI
 
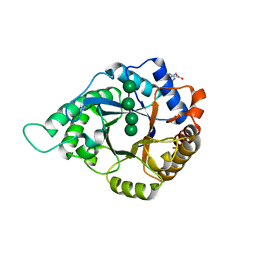 | | Complex structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanas, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
4UDW
 
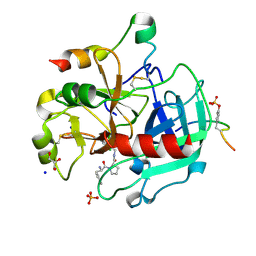 | | Thrombin in complex with 1-(2R)-2-amino-3-phenyl-propanoyl-N-(2, 5dichlorophenyl)methylpyrrolidine-2-carboxamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-(2,5-dichlorobenzyl)-L-prolinamide, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
4UE5
 
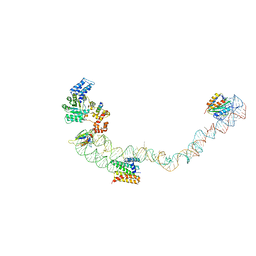 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5YM3
 
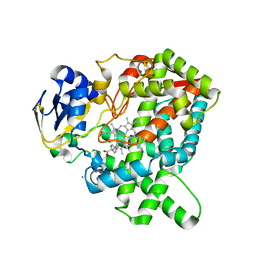 | | CYP76AH1-4pi from salvia miltiorrhiza | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Ferruginol synthase, MANGANESE (II) ION, ... | | Authors: | Chang, Z. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
4UFB
 
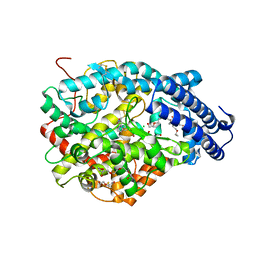 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with Lys-Pro | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ac-Sdkp Hydrolysis by Angiotensin-I Converting Enzyme
Sci.Rep., 5, 2015
|
|
5YN6
 
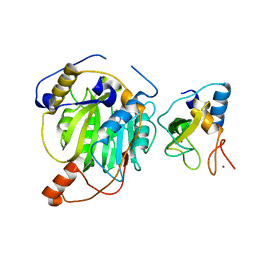 | | Crystal structure of MERS-CoV nsp10/nsp16 complex bound to SAM | | Descriptor: | S-ADENOSYLMETHIONINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, M06 | | Authors: | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | Deposit date: | 2014-05-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
5YNL
 
 | | Crystal structure of a cold-adapted arginase from psychrophilic yeast, Glaciozyma antarctica | | Descriptor: | Arginase | | Authors: | Yusof, N.Y, Quay, D.H.X, Illias, R.M, Firdaus-Raih, M, Abu-Bakar, F.D, Mahadi, N.M, Murad, A.M.A. | | Deposit date: | 2017-10-24 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Crystal structure of a cold-adapted arginase from psychrophilic yeast, Glaciozyma antarctica
To Be Published
|
|
4UN3
 
 | | Crystal structure of Cas9 bound to PAM-containing DNA target | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
5YNN
 
 | | Crystal structure of MERS-CoV nsp16/nsp10complex bound to sinefungin and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
4UQK
 
 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4KD3
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A/V66A/I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
5YO2
 
 | | The crystal structure of Rv2747 from Mycobacterium tuberculosis in complex with Acetyl CoA and L-Arginine | | Descriptor: | ACETYL COENZYME *A, ARGININE, Amino-acid acetyltransferase | | Authors: | Singh, E, Tiruttani Subhramanyam, U.K, Pal, R.K, Srinivasan, A, Gourinath, S, Das, U. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural insights into the substrate binding mechanism of novel ArgA from Mycobacterium tuberculosis
Int. J. Biol. Macromol., 125, 2019
|
|
