1MY4
 
 | | crystal structure of glutamate receptor ligand-binding core in complex with iodo-willardiine in the Zn crystal form | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
4ZV6
 
 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
6EDS
 
 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Glucagon and Substrate-selective Macrocyclic Inhibitor 63 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R, Welsh, A.J. | | Deposit date: | 2018-08-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18071723 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
1W7Z
 
 | | Crystal structure of the free (uncomplexed) Ecballium elaterium trypsin inhibitor (EETI-II) | | Descriptor: | FORMIC ACID, SODIUM ION, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Debreczeni, J.E, Pape, T, Kolmar, H, Schneider, T.R, Uson, I, Scheldrick, G.M. | | Deposit date: | 2004-09-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7TJC
 
 | | VHH Chl-B2 in complex with Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, GLYCEROL, VHH-Chl-B2 | | Authors: | Nordeen, S.A, Schwartz, T.U. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and specificity of an anti-chloramphenicol single domain antibody for detection of amphenicol residues.
Protein Sci., 31, 2022
|
|
7S71
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI35 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S72
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI36 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6W
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6X
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI30 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6Y
 
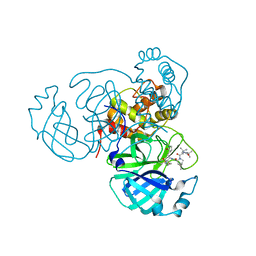 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI32 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S75
 
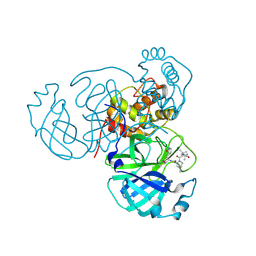 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI42 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S74
 
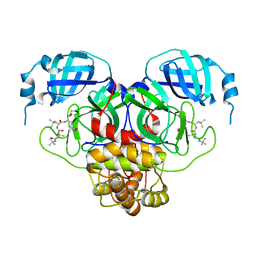 | |
7S6Z
 
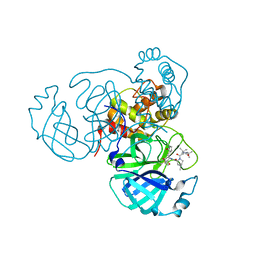 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S73
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI37 | | Descriptor: | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name), 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
8FTT
 
 | |
6RZX
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
7S8H
 
 | |
8TN8
 
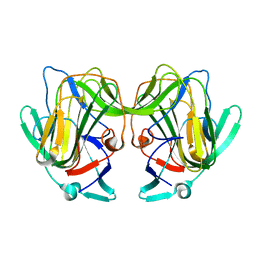 | |
5TMC
 
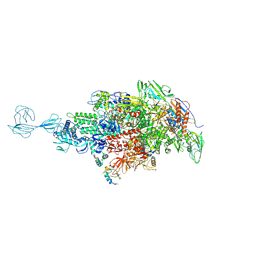 | | Re-refinement of Thermus thermopiles DNA-directed RNA polymerase structure | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Wang, J. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-23 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | On the validation of crystallographic symmetry and the quality of structures.
Protein Sci., 24, 2015
|
|
4GK6
 
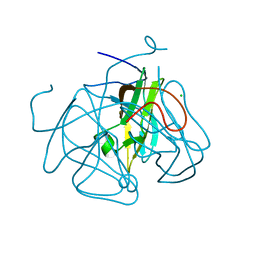 | |
6DM8
 
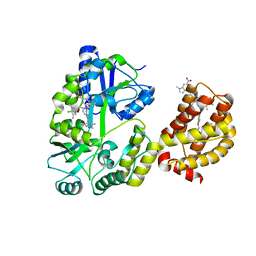 | | Understanding the Species Selectivity of Myeloid cell leukemia-1 (Mcl-1) inhibitors | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog - MBP chimera, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding the Species Selectivity of Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors.
Biochemistry, 57, 2018
|
|
1N0X
 
 | | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, B2.1 peptide, GLYCEROL, ... | | Authors: | Saphire, E.O, Montero, M, Menendez, A, Irving, M.B, Zwick, M.B, Parren, P.W.H.I, Burton, D.R, Scott, J.K, Wilson, I.A. | | Deposit date: | 2002-10-15 | | Release date: | 2004-04-13 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope
To be Published
|
|
8PQ4
 
 | |
2UUJ
 
 | | Thrombin-hirugen-gw473178 ternary complex at 1.32A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-03 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UU8
 
 | | X-ray structure of Ni, Ca concanavalin A at Ultra-high resolution (0. 94A) | | Descriptor: | CALCIUM ION, CONCANAVALIN, NICKEL (II) ION | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-01 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
