1BOZ
 
 | | STRUCTURE-BASED DESIGN AND SYNTHESIS OF LIPOPHILIC 2,4-DIAMINO-6-SUBSTITUTED QUINAZOLINES AND THEIR EVALUATION AS INHIBITORS OF DIHYDROFOLATE REDUCTASE AND POTENTIAL ANTITUMOR AGENTS | | Descriptor: | N6-(2,5-DIMETHOXY-BENZYL)-N6-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (DIHYDROFOLATE REDUCTASE) | | Authors: | Gangjee, A, Vidwans, A.P, Vasudevan, A, Queener, S.F, Kisliuk, R.L, Cody, V, Li, R, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation as inhibitors of dihydrofolate reductases and potential antitumor agents.
J.Med.Chem., 41, 1998
|
|
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3NSQ
 
 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | Descriptor: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
8CH6
 
 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
8HCU
 
 | | Crystal structure of BCOR/PCGF1/KDM2B complex | | Descriptor: | ACETIC ACID, BCL-6 corepressor, Polycomb group RING finger protein 1, ... | | Authors: | Shen, F, Chen, R, Xu, J, Liu, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calcium modulates the tethering of BCOR-PRC1.1 enzymatic core to KDM2B via liquid-liquid phase separation.
Commun Biol, 7, 2024
|
|
1RVY
 
 | | E75Q MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE, COMPLEX WITH GLYCINE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-15 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1RV4
 
 | | E75L MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-12 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
6TWJ
 
 | |
6U4Y
 
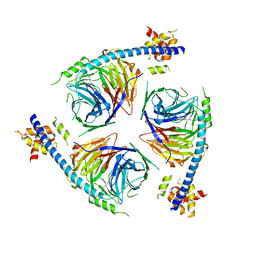 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TWM
 
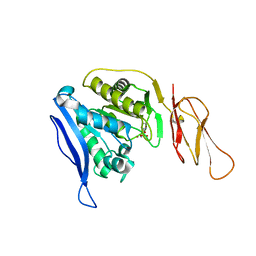 | | Product bound structure of the Ectoine utilization protein EutE (DoeB) from Ruegeria pomeroyi | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, ACETATE ION, N-acetyl-L-2,4-diaminobutyric acid deacetylase, ... | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
6TWK
 
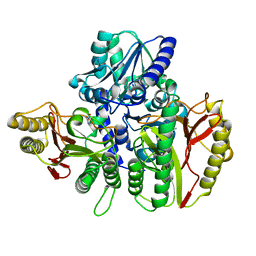 | | Substrate bound structure of the Ectoine utilization protein EutD (DoeA) from Halomonas elongata | | Descriptor: | (2~{R})-4-azanyl-2-[[(1~{S})-1-oxidanylethyl]amino]butanoic acid, (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Ectoine hydrolase DoeA | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
6TWL
 
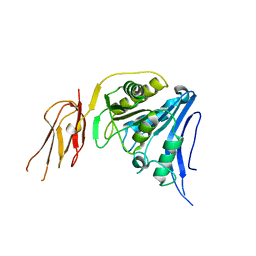 | |
2E1C
 
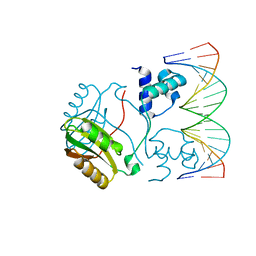 | | Structure of Putative HTH-type transcriptional regulator PH1519/DNA Complex | | Descriptor: | DNA (5'-D(*DAP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DT)-3'), Putative HTH-type transcriptional regulator PH1519 | | Authors: | Koike, H, Suzuki, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-12-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Feast/Famine Regulation by Transcription Factor FL11 for the Survival of the Hyperthermophilic Archaeon Pyrococcus OT3.
Structure, 15, 2007
|
|
4AIH
 
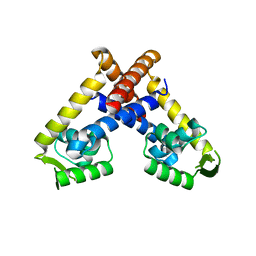 | | Crystal structure of RovA from Yersinia in its free form | | Descriptor: | TRANSCRIPTIONAL REGULATOR SLYA | | Authors: | Quade, N, Mendonca, C, Herbst, K, Heroven, A.K, Heinz, D.W, Dersch, P. | | Deposit date: | 2012-02-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Intrinsic Thermosensing by the Master Virulence Regulator Rova of Yersinia.
J.Biol.Chem., 287, 2012
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4AIK
 
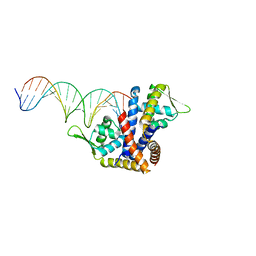 | | Crystal structure of RovA from Yersinia in complex with an invasin promoter fragment | | Descriptor: | ROVA PROMOTER FRAGMENT, 5'-D(*AP*TP*GP*AP*TP*AP*TP*TP *AP*TP**TP*TP*AP*TP*AP*TP*GP*AP*TP*AP*A)-3', 5'-D(*TP*TP*TP*AP*TP*CP*AP*TP *AP*TP*AP*AP*AP*TP*AP*AP*TP*AP*TP*CP*A)-3', ... | | Authors: | Quade, N, Mendonca, C, Herbst, K, Heroven, A.K, Heinz, D.W, Dersch, P. | | Deposit date: | 2012-02-10 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Intrinsic Thermosensing by the Master Virulence Regulator Rova of Yersinia.
J.Biol.Chem., 287, 2012
|
|
5HLG
 
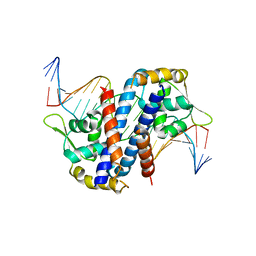 | | Structure of reduced AbfR bound to DNA | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*TP*CP*AP*AP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*TP*GP*AP*GP*T)-3'), MarR family transcriptional regulator | | Authors: | Liu, G, Liu, X, Gan, J, Yang, C.G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Redox-Sensing Mechanism of MarR-Type Regulator AbfR.
J. Am. Chem. Soc., 139, 2017
|
|
5HLH
 
 | | Crystal structure of the overoxidized AbfR bound to DNA | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*TP*CP*AP*AP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*TP*GP*AP*GP*T)-3'), MarR family transcriptional regulator | | Authors: | Liu, G, Liu, X, Gan, J, Yang, C.-G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Redox-Sensing Mechanism of MarR-Type Regulator AbfR.
J. Am. Chem. Soc., 139, 2017
|
|
2JGO
 
 | | Structure of the arsenated de novo designed peptide Coil Ser L9C | | Descriptor: | ARSENIC, COIL SER L9C, ZINC ION | | Authors: | Touw, D.S, Nordman, C.E, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2007-02-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identifying Important Structural Characteristics of Arsenic Resistance Proteins by Using Designed Three-Stranded Coiled Coils.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5F7V
 
 | |
4AIJ
 
 | | Crystal structure of RovA from Yersinia in complex with a rovA promoter fragment | | Descriptor: | ROVA PROMOTER FRAGMENT, 5'-D(*AP*AP*TP*TP*AP*TP*AP*TP *TP*AP*TP*TP*TP*GP*AP*AP*TP*TP*AP*AP*T)-3', 5'-D(*TP*AP*TP*TP*AP*AP*TP*TP *CP*AP*AP*AP*TP*AP*AP*TP*AP*TP*AP*AP*T)-3', ... | | Authors: | Quade, N, Mendonca, C, Herbst, K, Heroven, A.K, Heinz, D.W, Dersch, P. | | Deposit date: | 2012-02-10 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Intrinsic Thermosensing by the Master Virulence Regulator Rova of Yersinia.
J.Biol.Chem., 287, 2012
|
|
5DO8
 
 | | 1.8 Angstrom crystal structure of Listeria monocytogenes Lmo0184 alpha-1,6-glucosidase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Lmo0184 protein, ... | | Authors: | Light, S.H, Halavaty, A.S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-10 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure to function of an alpha-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis.
Nat Microbiol, 2, 2016
|
|
6G16
 
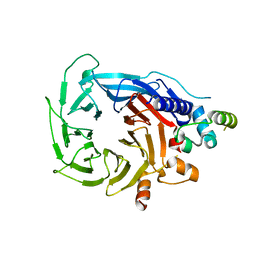 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
2JUP
 
 | | FBP28WW2 domain in complex with the PPLIPPPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-01 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2F41
 
 | |
