8V7F
 
 | | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T*())-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+
To Be Published
|
|
9EPM
 
 | | Mpro from SARS-CoV-2 with 4A mutation | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EWM
 
 | | Mpro from SARS-CoV-2 with R4Q R298Q double mutations | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EUS
 
 | | Mpro from SARS-CoV-2 with R298A mutation | | Descriptor: | GLYCEROL, Replicase polyprotein 1a | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8V7B
 
 | |
8V7H
 
 | |
8V7E
 
 | | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mg2+
To Be Published
|
|
9EUR
 
 | | Mpro WT from SARS-CoV-2 with 298Q mutation | | Descriptor: | Replicase polyprotein 1a | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EWN
 
 | | Mpro from SARS-CoV-2 with 4Q mutation | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8V7A
 
 | |
8V7I
 
 | |
8V7C
 
 | |
8V7K
 
 | |
8V7J
 
 | |
9EWO
 
 | | Mpro from SARS-CoV-2 with R4A R298A double mutations | | Descriptor: | Non-structural protein 11, SULFATE ION | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
2DQ3
 
 | |
3WCU
 
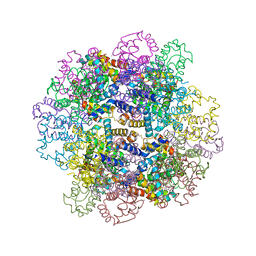 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Deoxygenated form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCV
 
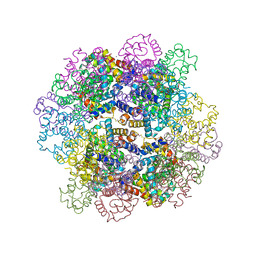 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: CA bound form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2M17
 
 | |
3WCW
 
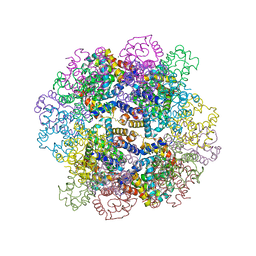 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: MG bound form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2UXA
 
 | | Crystal structure of the GluR2-flip ligand binding domain, r/g unedited. | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT GLUR2-FLIP, GLUTAMIC ACID, ZINC ION | | Authors: | Greger, I.H, Akamine, P, Khatri, L, Ziff, E.B. | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Developmentally Regulated, Combinatorial RNA Processing Modulates Ampa Receptor Biogenesis.
Neuron, 51, 2006
|
|
1U9B
 
 | | MURINE/HUMAN UBIQUITIN-CONJUGATING ENZYME UBC9 | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E9 | | Authors: | Tong, H, Hateboer, G, Perrakis, A, Bernards, R, Sixma, T.K. | | Deposit date: | 1997-05-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system.
J.Biol.Chem., 272, 1997
|
|
6CP9
 
 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | Descriptor: | CdiA, CdiI | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
3WCT
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Oxygenated form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2O43
 
 | | Structure of 23S rRNA of the large ribosomal subunit from Deinococcus radiodurans in complex with the macrolide erythromycylamine | | Descriptor: | (3R,4S,5S,6R,7R,9R,10S,11S,12R,13S,14R)-10-AMINO-6-{[(2S,3R,4S,6R)-4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PY RAN-2-YL]OXY}-14-ETHYL-7,12,13-TRIHYDROXY-4-{[(2R,4R,5S,6S)-5-HYDROXY-4-METHOXY-4,6-DIMETHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY }-3,5,7,9,11,13-HEXAMETHYLOXACYCLOTETRADECAN-2-ONE, 23S rRNA | | Authors: | Pyetan, E, Baram, D, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2006-12-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Chemical parameters influencing fine tuning in the binding of Macrolide antibiotics to the ribosomal tunnel
TO BE PUBLISHED
|
|
