3TKN
 
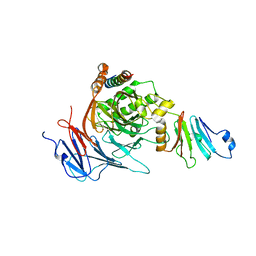 | | Structure of the Nup82-Nup159-Nup98 heterotrimer | | Descriptor: | Nucleoporin 98, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T.T, Hoelz, A. | | Deposit date: | 2011-08-28 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for the anchoring of proto-oncoprotein nup98 to the cytoplasmic face of the nuclear pore complex.
J.Mol.Biol., 419, 2012
|
|
3TLG
 
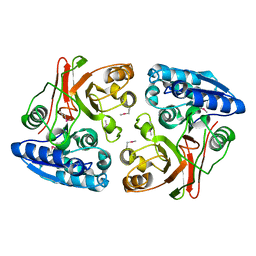 | |
3TKB
 
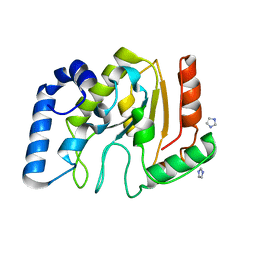 | | crystal structure of human uracil-DNA glycosylase D183G/K302R mutant | | Descriptor: | IMIDAZOLE, Uracil-DNA glycosylase | | Authors: | Assefa, N.G, Niiranen, L, Willassen, N.P, Smalas, A.O, Moe, E. | | Deposit date: | 2011-08-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thermal unfolding studies of cold adapted uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua). A comparative study with human UNG.
Comp.Biochem.Physiol. B: Biochem.Mol.Biol., 161, 2012
|
|
3TLU
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TZA
 
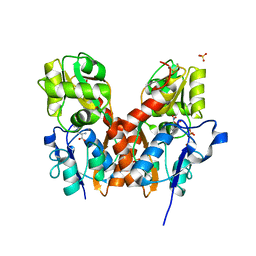 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid at 1.9A resolution | | Descriptor: | (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid, Glutamate receptor 2,Glutamate receptor 2, SULFATE ION | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new phenylalanine derivative acts as an antagonist at the AMPA receptor GluA2 and introduces partial domain closure: synthesis, resolution, pharmacology, and crystal structure
J.Med.Chem., 54, 2011
|
|
3TVZ
 
 | | Structure of Bacillus subtilis HmoB | | Descriptor: | Putative uncharacterized protein yhgC | | Authors: | Choe, J, Choi, S, Park, S. | | Deposit date: | 2011-09-21 | | Release date: | 2012-07-11 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus subtilis HmoB is a heme oxygenase with a novel structure.
Bmb Rep, 45, 2012
|
|
3U0C
 
 | |
3TYD
 
 | | Dihydropteroate Synthase in complex with PPi and DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, PYROPHOSPHATE 2-, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3TZG
 
 | |
3TZS
 
 | |
3U3E
 
 | |
3U3M
 
 | | Crystal structure of Human SULT1A1 bound to PAP and 3-Cyano-7-hydroxycoumarin | | Descriptor: | 7-hydroxy-2-oxo-2H-chromene-3-carbonitrile, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3U4S
 
 | | Histone Lysine demethylase JMJD2A in complex with T11C peptide substrate crosslinked to N-oxalyl-D-cysteine | | Descriptor: | HISTONE 3 TAIL ANALOG (T11C Peptide), Lysine-specific demethylase 4A, N-(carboxycarbonyl)-D-cysteine, ... | | Authors: | Ma, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Linking of 2-Oxoglutarate and Substrate Binding Sites Enables Potent and Highly Selective Inhibition of JmjC Histone Demethylases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3U64
 
 | | The Crystal Structure of Tat-T (Tp0956) | | Descriptor: | Protein TP_0956, SULFATE ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3U7C
 
 | | crystal structure of the V143I mutant of human carbonic anhydrase II | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | West, D.M, Kim, C.U, Robbins, A.H, Mckenna, R. | | Deposit date: | 2011-10-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | crystal structure of the V143I mutant of human carbonic anhydrase II
To be Published
|
|
3U7H
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTTp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U9U
 
 | | Crystal Structure of Extracellular Domain of Human ErbB4/Her4 in complex with the Fab fragment of mAb1479 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Hollmen, M, Liu, P, Wildiers, H, Reinvall, I, Vandorpe, T, Smeets, A, Deraedt, K, Vahlberg, T, Joensuu, H, Leahy, D.J, Schoffski, P, Elenius, K. | | Deposit date: | 2011-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Proteolytic processing of ErbB4 in breast cancer.
Plos One, 7, 2012
|
|
3U8H
 
 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | (S)-5-(4-BENZYLOXY-PHENYL)-4-(7-PHENYL-HEPTANOYLAMINO)-PENTANOIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
3UAV
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus | | Descriptor: | GLYCEROL, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TT6
 
 | |
3TXM
 
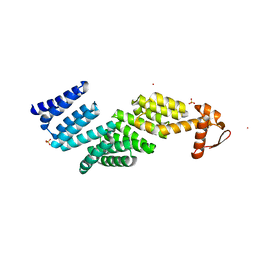 | |
3TTX
 
 | | Structure of the F413K variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3TWF
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3a | | Descriptor: | ACETYL GROUP, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TZC
 
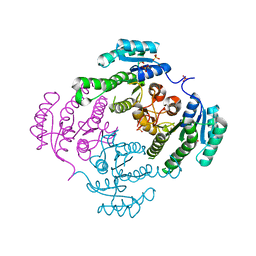 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(Y155F) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Domagalski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3U00
 
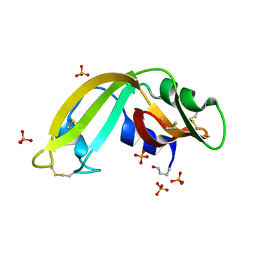 | | Crystal structure of wild-type onconase at 1.65 A resolution | | Descriptor: | PHOSPHATE ION, Protein P-30 | | Authors: | Kurpiewska, K, Torrent, G, Ribo, M, Vilanova, M, Loch, J, Lewinski, K. | | Deposit date: | 2011-09-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of wild-type onconase at 1.65 A resolution
To be Published
|
|
