4GOP
 
 | |
1JB7
 
 | |
4QTK
 
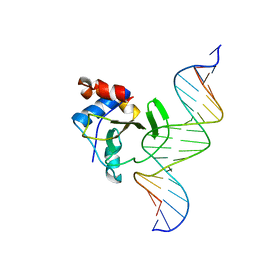 | | Complex of WOPR domain of Wor1 in Candida albicans with the 17bp dsDNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*TP*AP*AP*AP*CP*TP*TP*TP*TP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*AP*AP*AP*AP*AP*GP*TP*TP*TP*AP*AP*CP*TP*T)-3'), White-opaque regulator 1 | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of the WOPR-DNA complex and implications for Wor1 function in white-opaque switching of Candida albicans.
Cell Res., 24, 2014
|
|
3K75
 
 | |
3WU1
 
 | | Crystal structure of the ETS1-RUNX1-DNA ternary complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3'), Protein C-ets-1, ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
8DR4
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) without NTD | | Descriptor: | DNA (5'-D(P*AP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR3
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR1
 
 | | Consensus closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR5
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
1HRY
 
 | | THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTID-DIMENSIONAL HETERONUCLEAR-EDITED AND-FILTERED NMR | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*GP*TP*GP*C)-3'), HUMAN SRY | | Authors: | Clore, G.M, Werner, M.H, Huth, J.R, Gronenborn, A.M. | | Deposit date: | 1995-05-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex.
Cell(Cambridge,Mass.), 81, 1995
|
|
131D
 
 | | THE LOW-TEMPERATURE CRYSTAL STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA REVEALS BINDING OF A SPERMINE MOLECULE IN THE MINOR GROOVE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SODIUM ION, SPERMINE | | Authors: | Bancroft, D, Williams, L.D, Rich, A, Egli, M. | | Deposit date: | 1993-06-18 | | Release date: | 1993-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove.
Biochemistry, 33, 1994
|
|
145D
 
 | |
1A31
 
 | | HUMAN RECONSTITUTED DNA TOPOISOMERASE I IN COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*5IUP*5IU*TP*GP*AP*AP*AP*AP*AP*5IUP*5IUP*5IUP*5IUP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*5IUP*5IUP*5IUP*5IUP*CP*AP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), PROTEIN (TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-27 | | Release date: | 1998-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1WID
 
 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | Descriptor: | DNA-binding protein RAV1 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
181D
 
 | |
180D
 
 | |
187D
 
 | |
3E54
 
 | | Archaeal Intron-encoded Homing Endonuclease I-Vdi141I Complexed With DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DGP*DAP*DCP*DTP*DCP*DTP*DCP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DGP*DGP*DCP*DTP*DAP*DCP*DCP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(P*DGP*DAP*DGP*DAP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Nomura, N, Nomura, Y, Sussman, D, Stoddard, B.L. | | Deposit date: | 2008-08-13 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of a common rDNA target site in archaea and eukarya by analogous LAGLIDADG and His-Cys box homing endonucleases
Nucleic Acids Res., 36, 2008
|
|
1A35
 
 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
5LEW
 
 | | DNA polymerase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA polymerase III subunit alpha, SULFATE ION, ... | | Authors: | Banos-Mateos, S, Lang, U.F, Maslen, S.L, Skehel, J.M, Lamers, M.H. | | Deposit date: | 2016-06-30 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-fidelity DNA replication in Mycobacterium tuberculosis relies on a trinuclear zinc center.
Nat Commun, 8, 2017
|
|
1APL
 
 | | CRYSTAL STRUCTURE OF A MAT-ALPHA2 HOMEODOMAIN-OPERATOR COMPLEX SUGGESTS A GENERAL MODEL FOR HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*C P*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT-ALPHA2 HOMEODOMAIN) | | Authors: | Wolberger, C, Vershon, A.K, Liu, B, Johnson, A.D, Pabo, C.O. | | Deposit date: | 1993-10-04 | | Release date: | 1993-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 67, 1991
|
|
1BNA
 
 | | STRUCTURE OF A B-DNA DODECAMER. CONFORMATION AND DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Drew, H.R, Wing, R.M, Takano, T, Broka, C, Tanaka, S, Itakura, K, Dickerson, R.E. | | Deposit date: | 1981-01-26 | | Release date: | 1981-05-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a B-DNA dodecamer: conformation and dynamics.
Proc.Natl.Acad.Sci.USA, 78, 1981
|
|
9EOA
 
 | | Cryo_EM structure of human FAN1 in complex with 5' flap DNA substrate and PCNA | | Descriptor: | DNA (continuous), DNA (post-nick), DNA (pre-nick), ... | | Authors: | Jeyasankar, G, Salerno-Kochan, A, Thomsen, M. | | Deposit date: | 2024-03-14 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | A FAN1 point mutation associated with accelerated Huntington's disease progression alters its PCNA-mediated assembly on DNA.
Nat Commun, 16, 2025
|
|
2LEX
 
 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
8DQZ
 
 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
