3LMU
 
 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LC3
 
 | | Benzothiophene Inhibitors of Factor IXa | | Descriptor: | 1-[5-(3,4-dimethoxyphenyl)-1-benzothiophen-2-yl]methanediamine, CALCIUM ION, Coagulation factor IX | | Authors: | Wang, S, Beck, R. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Studies of Benzothiophene Template as Potent Factor IXa (FIXa) Inhibitors in Thrombosis.
J.Med.Chem., 53, 2010
|
|
3L7B
 
 | | Crystal Structure of Glycogen Phosphorylase DK3 complex | | Descriptor: | 4-amino-1-(3-deoxy-3-fluoro-beta-D-glucopyranosyl)pyrimidin-2(1H)-one, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
3LPK
 
 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9B
 
 | | Crystal Structure of Rat Otoferlin C2A | | Descriptor: | MAGNESIUM ION, Otoferlin | | Authors: | Helfmann, S, Neumann, P. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the C2A domain of otoferlin reveals an unconventional top loop region.
J.Mol.Biol., 406, 2011
|
|
3LRS
 
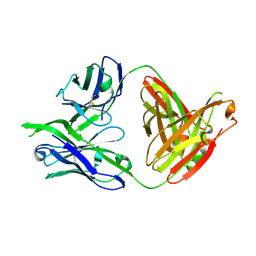 | | Structure of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PG-16 Heavy Chain Fab, PG-16 Light Chain Fab | | Authors: | Pancera, M, Kwong, P.D. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
3LSQ
 
 | |
3L9Z
 
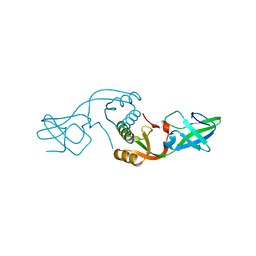 | | Crystal Structure of UreE from Helicobacter pylori (apo form) | | Descriptor: | Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3LAA
 
 | |
3LC2
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from methicillin resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-09 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LDI
 
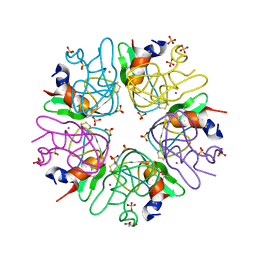 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | GLYCEROL, MERCURY (II) ION, Pancreatic trypsin inhibitor, ... | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding
Biochem.Biophys.Res.Commun., 397, 2010
|
|
3L6E
 
 | | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas hydrophila subsp. hydrophila ATCC 7966 | | Descriptor: | Oxidoreductase, short-chain dehydrogenase/reductase family, SULFATE ION | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-23 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas
hydrophila subsp. hydrophila ATCC 7966
To be Published
|
|
3L6T
 
 | |
3LHX
 
 | |
3LIE
 
 | |
3LF2
 
 | | NADPH Bound Structure of the Short Chain Oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 Containing an Atypical Catalytic Center | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Huether, R, Umland, T.C, Duax, W.L. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The short-chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center.
Protein Sci., 19, 2010
|
|
3LJ1
 
 | | IRE1 complexed with Cdk1/2 Inhibitor III | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lee, K.P.K, Sicheri, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1.
Mol.Cell, 38, 2010
|
|
3LK5
 
 | | Crystal structure of putative Geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum | | Descriptor: | GLYCEROL, Geranylgeranyl pyrophosphate synthase | | Authors: | Malashkevich, V.N, Toro, R, Patskovsky, Y, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative Geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum Atcc 13032
To be Published
|
|
3LGX
 
 | |
3LKX
 
 | | Human nac dimerization domain | | Descriptor: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | Authors: | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
3LL1
 
 | |
3LLC
 
 | |
3LM2
 
 | |
3LME
 
 | |
3LN3
 
 | |
