4JZV
 
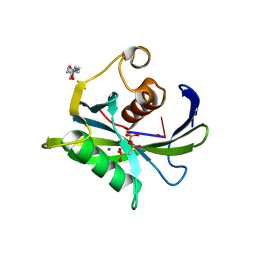 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZS
 
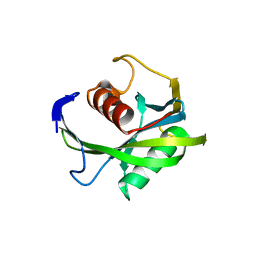 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZU
 
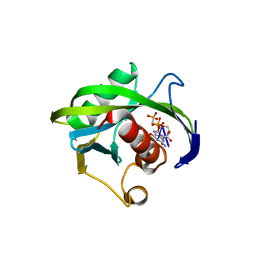 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - first guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RNA (5'-R(*(GCP)P*G)-3'), RNA PYROPHOSPHOHYDROLASE | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3A6V
 
 | |
4KG4
 
 | |
3A6T
 
 | | Crystal structure of MutT-8-OXO-DGMP complex | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, Mutator mutT protein, SODIUM ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3ACA
 
 | |
2RRK
 
 | |
3A6U
 
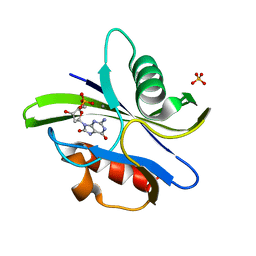 | | Crystal structure of MutT-8-OXO-dGMP-MN(II) complex | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mutator mutT protein, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3A6S
 
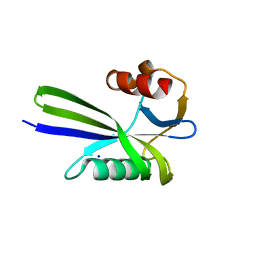 | | Crystal structure of the MutT protein | | Descriptor: | L(+)-TARTARIC ACID, Mutator mutT protein, SODIUM ION | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
4K6E
 
 | |
3AC9
 
 | |
4KG3
 
 | |
4KYX
 
 | |
2VNP
 
 | | Monoclinic form of IDI-1 | | Descriptor: | 2-DIMETHYLAMINO-ETHYL-DIPHOSPHATE, ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE, MAGNESIUM ION, ... | | Authors: | de Ruyck, J, Oudjama, Y, Wouters, J. | | Deposit date: | 2008-02-06 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Monoclinic Form of Isopentenyl Diphosphate Isomerase: A Case of Polymorphism in Biomolecular Crystals.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2W4E
 
 | | Structure of an N-terminally truncated Nudix hydrolase DR2204 from Deinococcus radiodurans | | Descriptor: | MUTT/NUDIX FAMILY PROTEIN | | Authors: | Goncalves, A.M.D, Fioravanti, E, Stelter, M, McSweeney, S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an N-Terminally Truncated Nudix Hydrolase Dr2204 from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3BM4
 
 | | Crystal Structure of Human ADP-ribose Pyrophosphatase NUDT5 In complex with magnesium and AMPcpr | | Descriptor: | ADP-sugar pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MAGNESIUM ION | | Authors: | Zha, M, Guo, Q, Zhang, Y, Zhong, C, Ou, Y, Ding, J. | | Deposit date: | 2007-12-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of ADP-Ribose Hydrolysis By Human NUDT5 From Structural and Kinetic Studies
J.Mol.Biol., 379, 2008
|
|
7TN4
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 3-diphosphoinositol 1,2,4,5-tetrakisphosphate (3-PP-IP4), Mg and Fluoride ion | | Descriptor: | (1R,2S,3R,4R,5S,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2022-01-20 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and catalytic analyses of the InsP 6 kinase activities of higher plant ITPKs.
Faseb J., 36, 2022
|
|
3COU
 
 | | Crystal structure of human Nudix motif 16 (NUDT16) | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 16 | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Van Den Berg, S, Welin, M, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Nudix motif 16 (NUDT16).
To be Published
|
|
3DKU
 
 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
3E57
 
 | |
7WWA
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 2.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW7
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW8
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 5 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW9
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
