1IZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1 mutant enzyme w398v | | Descriptor: | CALCIUM ION, amylase | | Authors: | Ohtaki, A, Iguchi, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual conversion of substrate specificities of Thermoactinomyces vulgaris R-47 alpha-amylases TVAI and TVAII by site-directed mutagenesis
CARBOHYDR.RES., 338, 2003
|
|
2BY0
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-28 | | Release date: | 2006-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6JHG
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121
To Be Published
|
|
6JMQ
 
 | | LAT1-CD98hc complex bound to MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2BXZ
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-28 | | Release date: | 2006-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-03-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
1J0H
 
 | |
7P4W
 
 | |
2BHU
 
 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MALTOOLIGOSYLTREHALOSE TREHALOHYDROLASE, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-18 | | Release date: | 2005-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
6JFJ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1IV8
 
 | |
2BHZ
 
 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with maltose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
6JMR
 
 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1J0K
 
 | |
1J0I
 
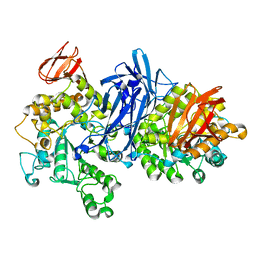 | | Crystal structure of neopullulanase complex with panose | | Descriptor: | alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, neopullulanase | | Authors: | Hondoh, H, Kuriki, T, Matsuura, Y. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase
J.Mol.Biol., 326, 2003
|
|
7P44
 
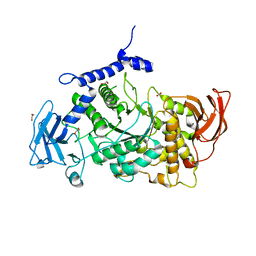 | | Structure of CgGBE in P21212 space group | | Descriptor: | 1,2-ETHANEDIOL, 1,4-alpha-glucan-branching enzyme | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7P45
 
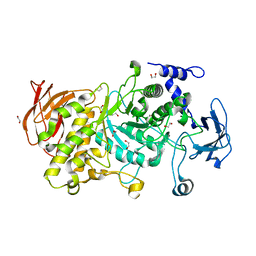 | | Structure of CgGBE in P212121 space group | | Descriptor: | 1,2-ETHANEDIOL, 1,4-alpha-glucan-branching enzyme | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7P43
 
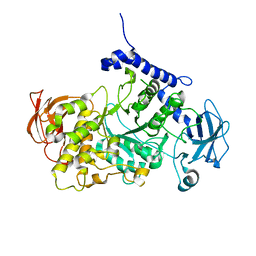 | | Structure of CgGBE in complex with maltotriose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7P9V
 
 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9U
 
 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
4MAZ
 
 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4MB1
 
 | | The Structure of MalL mutant enzyme G202P from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Oligo-1,6-glucosidase 1 | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4LQ1
 
 | |
5X7U
 
 | | Trehalose synthase from Thermobaculum terrenum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Trehalose synthase | | Authors: | Su, J, Wang, F. | | Deposit date: | 2017-02-27 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Characteristics and Function of a New Kind of Thermostable Trehalose Synthase from Thermobaculum terrenum.
J. Agric. Food Chem., 65, 2017
|
|
4M56
 
 | | The Structure of Wild-type MalL from Bacillus subtilis | | Descriptor: | D-glucose, GLYCEROL, Oligo-1,6-glucosidase 1, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
