2HCI
 
 | | Structure of Human Mip-3a Chemokine | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, SULFITE ION, ... | | Authors: | Malik, Z.A, Tack, B.F. | | Deposit date: | 2006-06-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of human MIP-3alpha chemokine.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2IL8
 
 | |
2JYO
 
 | | NMR Solution structure of Human MIP-3alpha/CCL20 | | Descriptor: | C-C motif chemokine 20 (Small-inducible cytokine A20) (Macrophage inflammatory protein 3 alpha) (MIP-3-alpha) (Liver and activation-regulated chemokine) (CC chemokine LARC) (Beta chemokine exodus-1) | | Authors: | Chan, D.I, Hunter, H.N, Tack, B.F, Vogel, H.J. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Human macrophage inflammatory protein 3alpha: protein and peptide nuclear magnetic resonance solution structures, dimerization, dynamics, and anti-infective properties.
Antimicrob.Agents Chemother., 52, 2008
|
|
2K01
 
 | |
2JP1
 
 | |
1NAP
 
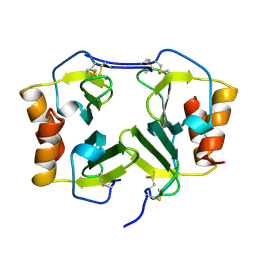 | |
1O80
 
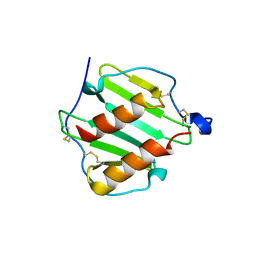 | |
1LV9
 
 | | CXCR3 Binding Chemokine IP-10/CXCL10 | | Descriptor: | Small inducible cytokine B10 | | Authors: | Booth, V, Keizer, D.W, Kamphuis, M.B, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2002-05-27 | | Release date: | 2002-09-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The CXCR3 binding chemokine IP-10/CXCL10: structure and receptor interactions.
Biochemistry, 41, 2002
|
|
1MI2
 
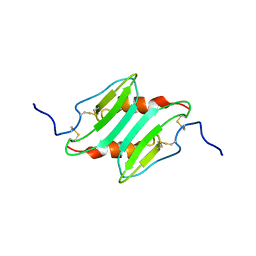 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
1O7Z
 
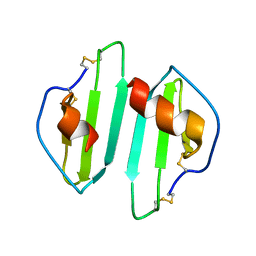 | |
1MSG
 
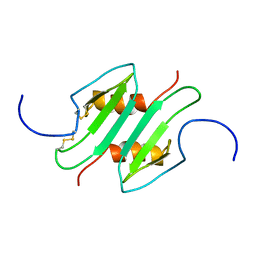 | |
1O7Y
 
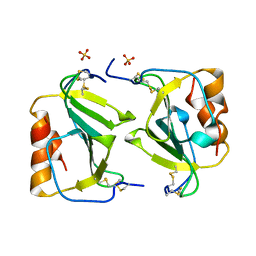 | | Crystal structure of IP-10 M-form | | Descriptor: | SMALL INDUCIBLE CYTOKINE B10, SULFATE ION | | Authors: | Swaminathan, G.J, Holloway, D.E, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-11-20 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Oligomeric Forms of the Ip-10/Cxcl10 Chemokine
Structure, 11, 2003
|
|
1NR4
 
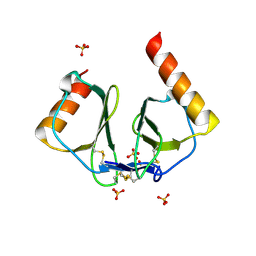 | | High resolution crystal structures of thymus and activation-regulated chemokine | | Descriptor: | SULFATE ION, Thymus and activation-regulated chemokine | | Authors: | Asojo, O.A, Boulegue, C, Hoover, D.M, Lu, W, Lubkowski, J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of thymus and activation-regulated chemokine (TARC).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NR2
 
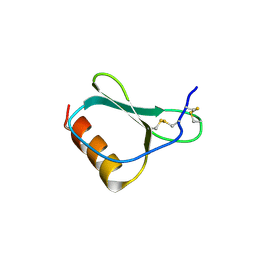 | | High resolution crystal structures of thymus and activation-regulated chemokine | | Descriptor: | Thymus and activation-regulated chemokine | | Authors: | Asojo, O.A, Boulegue, C, Hoover, D.M, Lu, W, Lubkowski, J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of thymus and activation-regulated chemokine (TARC).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1M8A
 
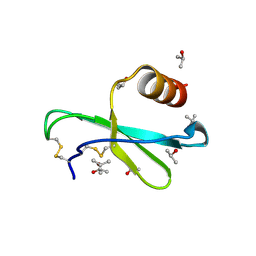 | | Human MIP-3alpha/CCL20 | | Descriptor: | ISOPROPYL ALCOHOL, Small inducible cytokine A20 | | Authors: | Hoover, D.M, Boulegue, C, Yang, D, Oppenheim, J.J, Tucker, K, Lu, W, Lubkowski, J. | | Deposit date: | 2002-07-24 | | Release date: | 2002-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human macrophage inflammatory protein-3alpha /CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins
J.Biol.Chem., 277, 2002
|
|
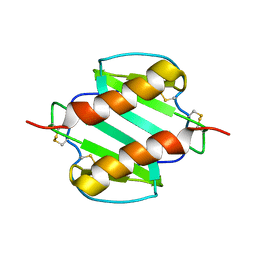
1MGS
 
 | |
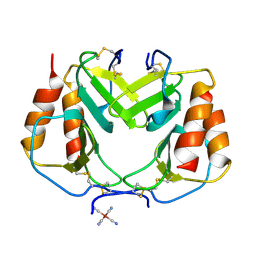
1PLF
 
 | |
1MSH
 
 | |
1NCV
 
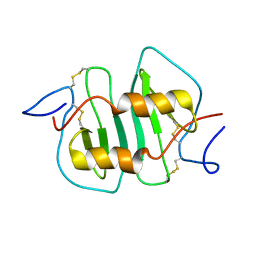 | | DETERMINATION CC-CHEMOKINE MCP-3, NMR, 7 STRUCTURES | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 3 | | Authors: | Meunier, S, Bernassau, J.M, Guillemot, J.C, Ferrara, P, Darbon, H. | | Deposit date: | 1997-02-05 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of CC chemokine monocyte chemoattractant protein 3 by 1H two-dimensional NMR spectroscopy.
Biochemistry, 36, 1997
|
|
1PFN
 
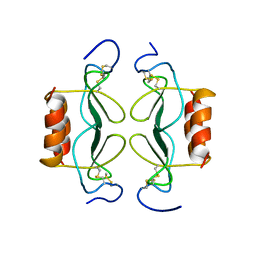 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1PFM
 
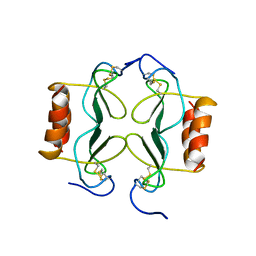 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1BO0
 
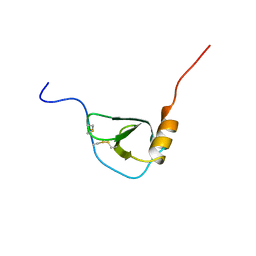 | | MONOCYTE CHEMOATTRACTANT PROTEIN-3, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (MONOCYTE CHEMOATTRACTANT PROTEIN-3) | | Authors: | Kwon, D, Lee, D, Sykes, B.D, Kim, K.-S. | | Deposit date: | 1998-08-10 | | Release date: | 1999-10-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a monomeric chemokine: monocyte chemoattractant protein-3.
FEBS Lett., 395, 1996
|
|
1EQT
 
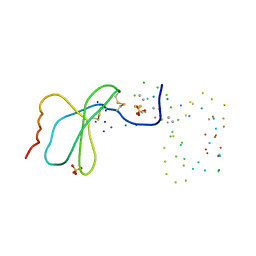 | | MET-RANTES | | Descriptor: | SULFATE ION, T-CELL SPECIFIC RANTES PROTEIN | | Authors: | Hoover, D.M, Shaw, J, Gryczynski, Z, Proudfoot, A.E.I, Wells, T. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of MET-RANTES: Comparison with Native RANTES and AOP-RANTES
PROTEIN PEPT.LETT., 7, 2000
|
|
1DOM
 
 | |
1DOL
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, I-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1 | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A. | | Deposit date: | 1996-11-22 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
